Credit: Nina Mars, University of Helsinki
In women, breast cancer is the most commonly diagnosed cancer and the leading cause of cancer-related deaths. Genetic predisposition is one of the key risk factors associated with the disease.
A study coordinated by University of Helsinki researchers examined the risk of developing the disease on both the population level and in individual carriers of specific gene defects which significantly increase breast cancer risk. The results, published today in the journal Nature Communications, are based on the FinnGen project encompassing more than 120,000 women, which combines genomic information with health data from national health registries.
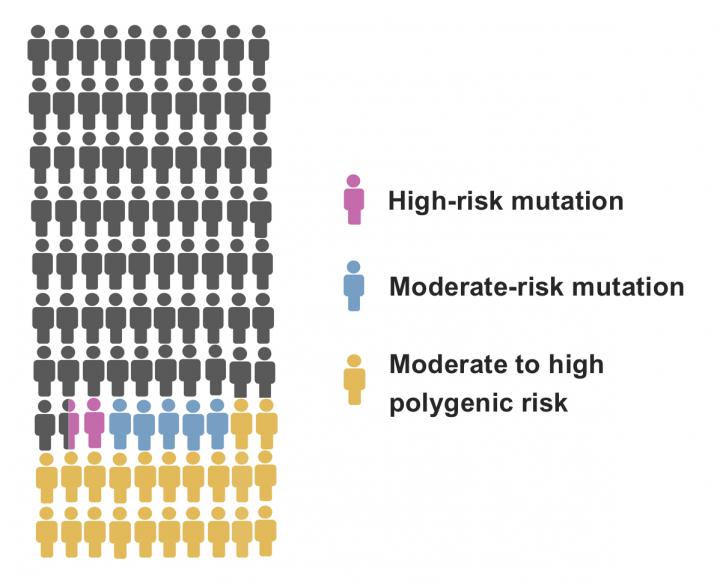
The study focused on specific mutations in the PALB2 and CHEK2 genes, the two most common high-risk breast cancer mutations in the Finnish population. A gene test for identifying these mutations is employed in the clinic when a strong hereditary predisposition to breast cancer is suspected in the family. The PALB2 mutation is found in approximately 1% of Finnish breast cancer patients, while the CHEK2 mutation is found in roughly 2.5%.
However, it has been known for a long time that these genes defects do not result in cancer in all women. The new study demonstrates that the lifetime risk of developing breast cancer among carriers of the mutations varies considerably on the basis of the cumulative effects of risk factors originating elsewhere in the genome.
The researchers determined for the individuals what is known as a polygenic risk score, which sums up a number of genetic risk factors associated with breast cancer risk. Individually, such genetic risk factors have a modest impact on the risk of developing the disease but, collectively pooled into a risk score, women with an unusually high or low risk of developing breast cancer can be distinguished from others.
“For carriers of either the PALB2 or CHEK2 mutation with an elevated polygenic risk, the lifetime risk of developing breast cancer may be as high as 60-80%. Correspondingly, low polygenic risk can compensate the impact of the mutation,” says Nina Mars, MD, from the University of Helsinki’s Institute for Molecular Medicine Finland (FIMM), who carried out the study.
“For CHEK2 carriers, for example, low polygenic risk score means that their risk does not differ from that of the overall population. A more accurate risk assessment such as this for carriers of significant pathogenic mutations would help target, for example, screening for breast cancer in order to identify cases in the early stage.”
The polygenic risk score proved to be a significant risk factor also on its own, as the researchers found that those in the top ten percent according to the score have at least as high a risk of developing the disease as carriers of the CHEK2 mutation. In particular, the polygenic risk score can provide a much more accurate disease risk assessment among the patient’s close relatives.
“Even in the case of marked accumulation of breast cancer cases in individual families, clinical gene panel testing identifies a high-risk mutation in only one out of every six patients. In fact, an exceptionally high polygenic risk score, with similar impact, underlies the breast cancer risk of many families. This is why risk-related information could be used especially in assessing the risk of patients’ close relatives,” says Professor Samuli Ripatti from the University of Helsinki, the principal investigator of the study.
The research group considers the utilisation of polygenic risk information a promising tool on the path towards an increasingly comprehensive risk assessment for breast cancer.
“Next we are starting the first trials where these scores will be integrated into the care of breast cancer patients and their families,” Ripatti adds.
“These exciting results chart a clear path towards significantly improved risk assessment that will ultimately save lives and reduce healthcare costs by ensuring that cancer screening is delivered to those who need it most,” says Mark Daly, the Director of FIMM.
###
Media Contact
Samuli Ripatti, professor
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




