Researchers have isolated a new strain of marine bacteria with unique characteristics from the ocean seabed.
Credit: Rikuan Zheng
Researchers have isolated a new strain of marine bacteria with unique characteristics from the ocean seabed.
The research, published today as a Reviewed Preprint in eLife, is described by the editors as an important study that advances our understanding of physiological mechanisms in deep-sea Planctomycetes bacteria, revealing unique characteristics such as being the only known species in the class of Phycisphaerae bacteria that uses a distinct budding model of division. It provides what the editors also say is convincing evidence that the new species is extensively involved in nitrogen assimilation and lives with a chronic virus (bacteriophage) that facilitates nitrogen metabolism. Nitrogen cycling by bacteria is an essential process that frees up nitrogen for building into nucleic acids, amino acids and proteins – the building blocks of life.
“Until recently, most research on the Planctomycetes family of bacteria has focused on strains in freshwater and shallow ocean environments, because of the logistical difficulties associated with sampling and cultivating deep-sea strains,” says lead author Rikuan Zheng, a research associate at the Institute of Oceanology, Chinese Academy of Sciences, Beijing, China, and the National Laboratory for Marine Science and Technology, Qingdao, China. “Most Planctomycetes bacteria have been isolated using growth media that are nutritionally poor, so we wanted to see if using a nutrient-rich medium would make it possible to culture and further characterise members of this poorly understood family.”
To isolate the novel bacterium, the team took sediment samples from a deep-sea cold seep, where Planctomycetes bacteria are known to reside, and then encouraged their growth by supplementing a standard growth medium with the antibiotic rifampicin and sources of nitrogen. They cultured these enriched bacteria on agar and evaluated individual colonies further by gene sequencing. Among the bacteria, they identified a strain called ZRK32 that grew faster than others, and looked likely to be a member of the genus Poriferisphaera. To confirm this, the team compared the genetic similarities between this strain and other members of the Poriferisphaera genus and found that it was distinguishable from Poriferisphaera corsica, the only other species with a valid published name. This suggests that ZRK32 is a novel species – which the team proposes to call Poriferisphaera hetertotrophicis.
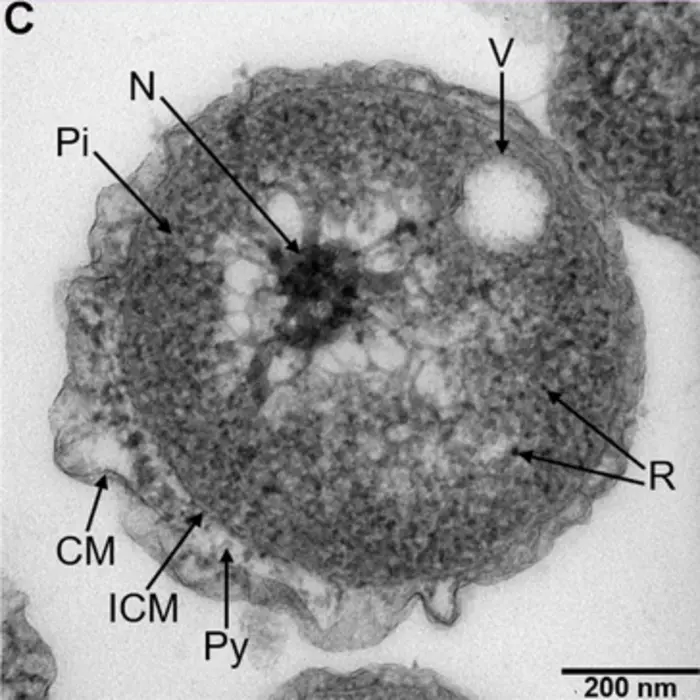
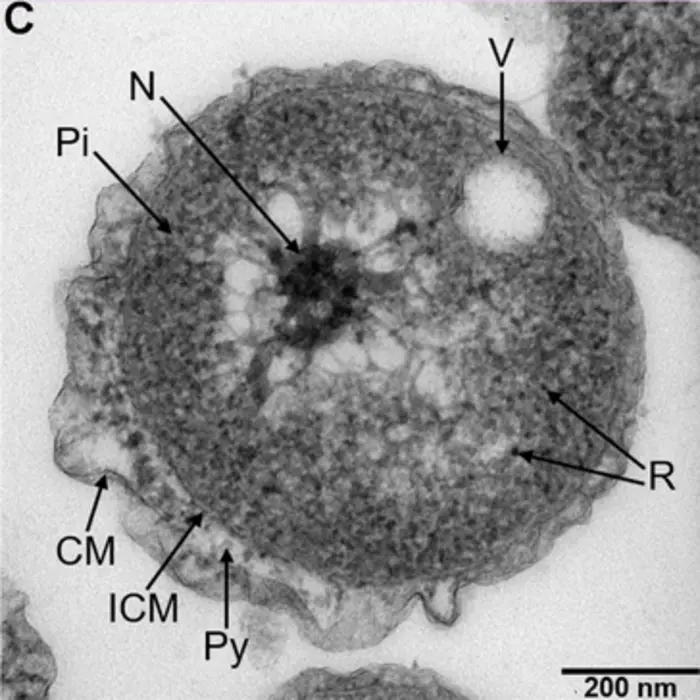
To learn more about this new species, the team studied its growth and how it multiplies. They found that, unlike other Planctomycetes family members, Poriferisphaera hetertotrophicis grows better in nutrient-rich media and multiplies via a budding mechanism, where parent cells create outgrowth buds that develop into daughter cells.
As the Planctomycetes bacteria family is known to play an important role in nitrogen cycling, the team next explored whether this was also the case for Poriferisphaera hetertotrophicis. To test this, they looked at the effects of different nitrogen-containing substances – nitrates, ammonia and nitrogen dioxide – on Poriferisphaera hetertotrophicis growth. They found that adding nitrogen in the form of a nitrate or ammonia increased growth, whereas adding it as a nitrite inhibited growth.
They also discovered that the addition of nitrate or ammonia caused the novel strain to release a bacteriophage – a type of virus that infects bacteria. Bacteriophages are widely distributed across oceans and can regulate nitrogen metabolism in their host bacteria. This bacteriophage – called phage-ZRK32 – was able to increase the growth of Poriferisphaera hetertotrophicis and other marine bacteria dramatically by facilitating nitrogen metabolism. Even though the team’s genetic analysis suggested Poriferisphaera hetertotrophicis contains all the necessary genes for metabolising nitrate and ammonia, chronic infection with this bacteriophage may help to further optimise nitrogen metabolism.
“Our analyses indicate that strain ZRK32 is a novel species, which grows best in nutrient-rich media and releases a bacteriophage in the presence of nitrogen,” concludes senior author Chaomin Sun, a Professor at the Institute of Oceanology, Chinese Academy of Sciences, and the National Laboratory for Marine Science and Technology. “This phage-ZRK32 is a chronic bacteriophage that lives within its host without killing it. Our findings provide a novel insight into nitrogen metabolism in Planctomycetes bacteria and a suitable model to study the interactions between Planctomycetes and viruses.”
##
Media contacts
Emily Packer, Media Relations Manager
eLife
01223 855373
George Litchfield, Marketing and PR Assistant
eLife
About eLife
eLife transforms research communication to create a future where a diverse, global community of scientists and researchers produces open and trusted results for the benefit of all. Independent, not-for-profit and supported by funders, we improve the way science is practised and shared. In support of our goal, we have launched a new publishing model that ends the accept/reject decision after peer review. Instead, papers invited for review will be published as a Reviewed Preprint that contains public peer reviews and an eLife assessment. We also continue to publish research that was accepted after peer review as part of our traditional process. eLife receives financial support and strategic guidance from the Howard Hughes Medical Institute, Knut and Alice Wallenberg Foundation, the Max Planck Society and Wellcome. Learn more at https://elifesciences.org/about.
To read the latest Microbiology and Infectious Disease research published in eLife, visit https://elifesciences.org/subjects/microbiology-infectious-disease.
Journal
eLife
DOI
10.7554/eLife.89874.1
Article Title
Physiological and metabolic insights into the first cultured anaerobic representative of deep-sea Planctomycetes bacteria
Article Publication Date
29-Aug-2023