Credit: UTSW
DALLAS – Dec. 9, 2019 – UT Southwestern researchers have developed a software tool that uses artificial intelligence to recognize cancer cells from digital pathology images – giving clinicians a powerful way of predicting patient outcomes.
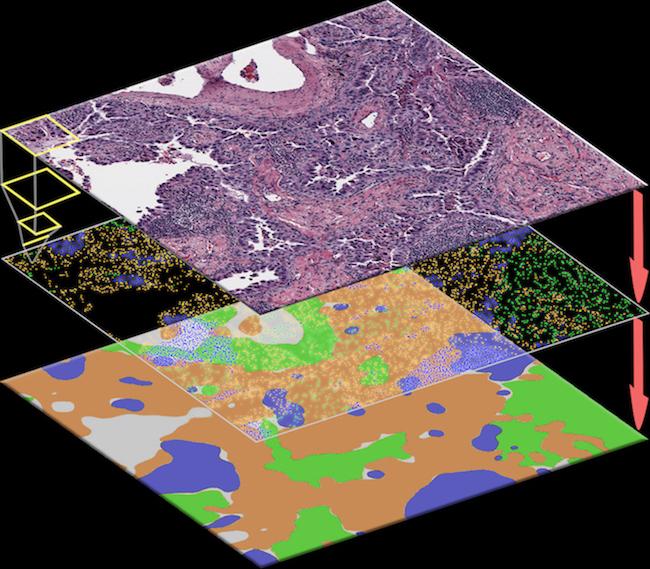
The spatial distribution of different types of cells can reveal a cancer’s growth pattern, its relationship with the surrounding microenvironment, and the body’s immune response. But the process of manually identifying all the cells in a pathology slide is extremely labor intensive and error-prone.
“As there are usually millions of cells in a tissue sample, a pathologist can only analyze so many slides in a day. To make a diagnosis, pathologists usually only examine several ‘representative’ regions in detail, rather than the whole slide. However, some important details could be missed by this approach,” said Dr. Guanghua “Andy” Xiao, corresponding author of a study published in EBioMedicine and Professor of Population and Data Sciences at UT Southwestern.
The human brain, Dr. Xiao added, is not good at picking up subtle morphological patterns. Therefore, a major technical challenge in systematically studying the tumor microenvironment is how to automatically classify different types of cells and quantify their spatial distributions, he said.
The AI algorithm that Dr. Xiao and his team developed, called ConvPath, overcomes these obstacles by using AI to classify cell types from lung cancer pathology images.
Here’s how it works: The ConvPath algorithm can “look” at cells and identify their types based on their appearance in the pathology images using an AI algorithm that learns from human pathologists. This algorithm effectively converts a pathology image into a “map” that displays the spatial distributions and interactions of tumor cells, stromal cells (i.e., the connective tissue cells), and lymphocytes (i.e., the white blood cells) in tumor tissue.
Whether tumor cells cluster well together or spread into stromal lymph nodes is a factor revealing the body’s immune response. So knowing that information can help doctors customize treatment plans and pinpoint the right immunotherapy.
Ultimately, the algorithm helps pathologists obtain the most accurate cancer cell analysis – in a much faster way.
“It is time-consuming and difficult for pathologists to locate very small tumor regions in tissue images, so this could greatly reduce the time that pathologists need to spend on each image,” said Dr. Xiao, who also has an appointment in the Lyda Hill Department of Bioinformatics and is a member of both the Quantitative Biomedical Research Center (QBRC) and the Harold C. Simmons Comprehensive Cancer Center at UTSW.
###
The ConvPath software – which incorporates image segmentation, deep learning, and feature extraction algorithms – is publicly accessible at https:/
The study’s lead authors include Shidan Wang, QBRC Data Scientist II; Dr. Tao Wang, Assistant Professor of Population and Data Sciences and in the Center for the Genetics of Host Defense; and Dr. Donghan M. Yang, QBRC Project Manager. Other co-authors from UT Southwestern include Dr. Yang Xie, Professor of Population and Data Sciences, a Professor in the Lyda Hill Department of Bioinformatics, and Director of the QBRC, and Dr. John Minna, Professor of Pharmacology and Internal Medicine and Director of the Hamon Center for Therapeutic Oncology Research. Dr. Minna holds the Sarah M. and Charles E. Seay Distinguished Chair in Cancer Research and the Max L. Thomas Distinguished Chair in Molecular Pulmonary Oncology.
The study was supported by the National Institutes of Health and the Cancer Prevention and Research Institute of Texas.
About UT Southwestern Medical Center
UT Southwestern, one of the premier academic medical centers in the nation, integrates pioneering biomedical research with exceptional clinical care and education. The institution’s faculty has received six Nobel Prizes, and includes 22 members of the National Academy of Sciences, 17 members of the National Academy of Medicine, and 15 Howard Hughes Medical Institute Investigators. The full-time faculty of more than 2,500 is responsible for groundbreaking medical advances and is committed to translating science-driven research quickly to new clinical treatments. UT Southwestern physicians provide care in about 80 specialties to more than 105,000 hospitalized patients, nearly 370,000 emergency room cases, and oversee approximately 3 million outpatient visits a year.
Media Contact
Deborah Wormser
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




