The genome is not a fixed code but flexible. It allows changes in the genes. Transposons, however, so-called jumping genes, interpret this flexibility in a much freer way than “normal” genes. They reproduce in the genome and chose their position themselves. Transposons can also jump into a gene and render it inoperative. Thus, they are an important distinguishing mark for the development of different organisms.
Unclear what triggers transposon activity
However, it is still unclear how jumping genes developed and what influences their activity. “In order to find out how, for instance, climate zones influence activity, we must be able to compare the frequency of transposons in different populations — in different groups of individuals,” explained bioinformatician Robert Kofler from the Institute of Population Genetics at the University of Veterinary Medicine, Vienna. But this frequency has not yet been determined precisely.
New software for a low-priced method
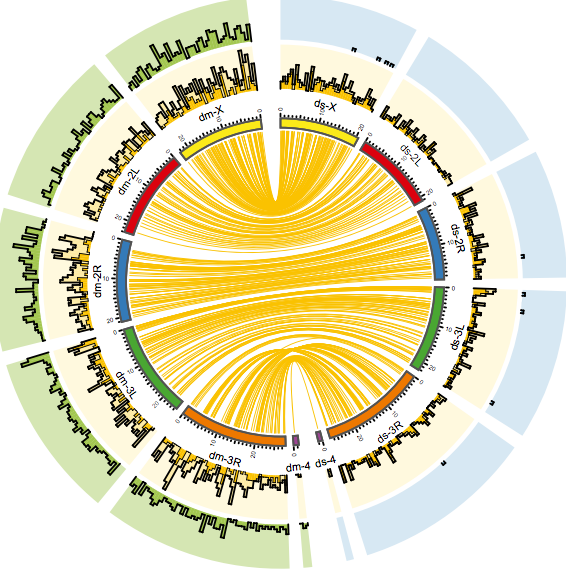
Transposons are detected by DNA sequencing. But this detection cannot be carried out for every single member of a population. “At the moment, this would go beyond the available resources regarding finance and amount of work. The only — and much cheaper — option is to analyse an entire population in one reaction,” explained last author Christian Schlötterer. This method, which he has established using the example of fruit flies, is called Pool-Seq. It is also routinely applied to detect transposons. Existing analysis programmes, however, could not provide a precise result in this case. So far, each analysis has been biased by different factors such as the sequencing depth and the distance between paired reads.
For this purpose, Kofler developed the new software PoPoolationTE2. “If we sequence entire populations, each reaction provides a different result. The number of mixed individuals is always the same, but the single individuals differ,” explained Kofler. Furthermore, technical differences in the sample processing, among others, have influenced the analysis so far. PoPoolationTE2 is not affected by these factors. Thus, questions about the activity of transposons can be answered precisely for Pool-Seq reactions.
Interesting for cancer research
“The unbiased detection of transposon abundance enables a low-price comparison of populations from, for instance, different climate zones. In a next step, we can find out if a transposon is very active in a particular climate zone,” said Kofler. In principle, the bioinformatician has developed this new software for Pool-Seq. But as this method is also applied in medical research and diagnosis, the programme is also interesting for cancer research or the detection of neurological changes since transposons also occur in the brain.
Lab experiments confirm influencing factors
Lab experiments can indicate the factors influencing transposons. Last author Schlötterer explained these factors referring to an experiment with fruit flies: “We breed a hundred generations per population and expose them to different stimuli. We sequence at every tenth generation and determine if a stimulus has influenced the activity of the transposons. Thus, we can describe the activity of transposons in fast motion, so to say.” If the abundance is low, the scientists assume that the transposons are only starting to become more frequent. If a transposon reproduces very quickly in a particular population, this is called an invasion. If a jumping gene is detected in an entire population and not in another one, it could have been positively selected.
Story Source:
The above post is reprinted from materials provided by Veterinärmedizinische Universität Wien.
Journal Reference:
Robert Kofler, Daniel Gómez-Sánchez, Christian Schlötterer. PoPoolationTE2: Comparative Population Genomics of Transposable Elements Using Pool-Seq. Molecular Biology and Evolution, 2016; msw137 DOI: 10.1093/molbev/msw137
The post New software helps to find out why ‘jumping genes’ are activated appeared first on Scienmag.