The proteins that make up our cells hold within an entire world of information, which, when unlocked, can give us insights into the origins of many essential biological phenomena. This information is gathered using an analytical technique known as “single-cell proteomics”, in which a single-cell analysis is performed to observe the characteristics of individual cells at their protein level. Over the years, scientists have used single-cell proteomics in the fields of cancer genomics, cell differentiation, and tissue development. However, current proteomics techniques suffer from low recovery rate of protein samples, low throughput, and lack of versatility.
Credit: Takeshi Masuda from Kumamoto University
The proteins that make up our cells hold within an entire world of information, which, when unlocked, can give us insights into the origins of many essential biological phenomena. This information is gathered using an analytical technique known as “single-cell proteomics”, in which a single-cell analysis is performed to observe the characteristics of individual cells at their protein level. Over the years, scientists have used single-cell proteomics in the fields of cancer genomics, cell differentiation, and tissue development. However, current proteomics techniques suffer from low recovery rate of protein samples, low throughput, and lack of versatility.
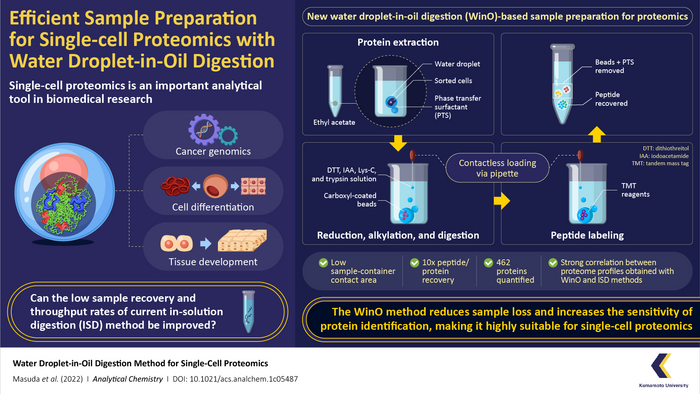
Fortunately, a team of researchers from Japan and USA led by Assistant Professor Takeshi Masuda from Kumamoto University in Japan have found a solution to these problems. In a recent study made available online on 11 July 2022 and published in Volume 94, Issue 29 of Analytical Chemistry on 26 July 2022, the team introduced a simple yet highly efficient sample preparation method for single-cell proteomics called “water droplet-in-oil method” (WinO). The technique uses the immiscibility of water with oil/organic solvent to its advantage to prepare protein samples with minimum loss and increased chances of sample recovery.
“To make single cell-proteomics more efficient, we either need to amplify the protein sample or make sure none of it is lost during sample preparation. Since we didn’t have the means to do the former, it was crucial that we reduced absorption losses during sample preparation steps like sample transfer,” explains Dr. Masuda. “The WinO technique not only reduces sample loss through adsorption but also provides better throughput when compared with conventional methods.”
For the WinO process, the team first prepared an extraction buffer by mixing one microliter of water with phase transfer surfactants (which increase the solubility of hydrophobic proteins) and hydrophobic carboxyl-coated nanomagnetic beads. This mixture was then dropped into 50 microliters of ethyl acetate.
The next step was protein extraction, which was performed by adding cell droplets from the cell sorter to the ethyl acetate-water droplet combo and spinning it in a centrifuge to allow the protein to accumulate within the water droplet. After the extraction, the sample was digested using a protein enzyme, Lys-C, and labeled using a “tandem mass tag” reagent . The extracted-digested-labeled sample was then purified and recovered for single-cell analysis and proteomic profiles.
To compare the efficacy of the WinO method against conventional methods, the team also prepared samples using the standard in-solution digestion (ISD) method and carried out proteomic analysis. They found that the WinO method led to a 10-fold increase in protein and peptide recovery compared to ISD. This remarkable improvement was attributed to a reduced contact area between the extraction solution and the sample container.
To analyze the sensitivity of both methods, the team also compared the obtained proteomic profiles. They observed a high correlation between proteomic profiles obtained for 100 cells using WinO and that for 10,000 cells using ISD. Furthermore, the team successfully quantified 462 proteins using WinO, demonstrating that it provided a much higher throughput and extraction efficiency than conventional techniques.
The enhanced protein recovery and identification ability provided by WinO could enable a closer look at the protein expression of cancer cells and a better understanding of the mechanisms underlying anticancer drug resistance. Further, WinO can be semi-automated using a liquid handling robot, making it suitable for high-speed, large-capacity processing of samples. “Our research could allow scientists to perform proteomics on rare and limited sample amounts as well as provide a novel perspective on protein expression, opening up possibilities for discovering new biological phenomena,” concludes Dr. Masuda.
We certainly hope his visions are realized soon!
About Assiatant Professor Takeshi Masuda
Dr. Takeshi Masuda is an Assistant Professor at the Faculty of Life Sciences at Kumamoto University, Japan. He worked as a project research assistant and research associate at the Institute of Advanced Biosciences in Keio University, Japan from 2004 to 2013. Prior to his appointment at the university in 2016, he was a postdoctoral researcher at the University of Washington and Harvard Medical School in USA. His current research interests include single-cell analysis, proteomics, biological mass spectrometry, and advanced sample preparation techniques.
Journal
Analytical Chemistry
DOI
10.1021/acs.analchem.1c05487
Method of Research
Experimental study
Subject of Research
Cells
Article Title
Water Droplet-in-Oil Digestion Method for Single-Cell Proteomics
Article Publication Date
26-Jul-2022
COI Statement
The authors declare no competing financial interest




