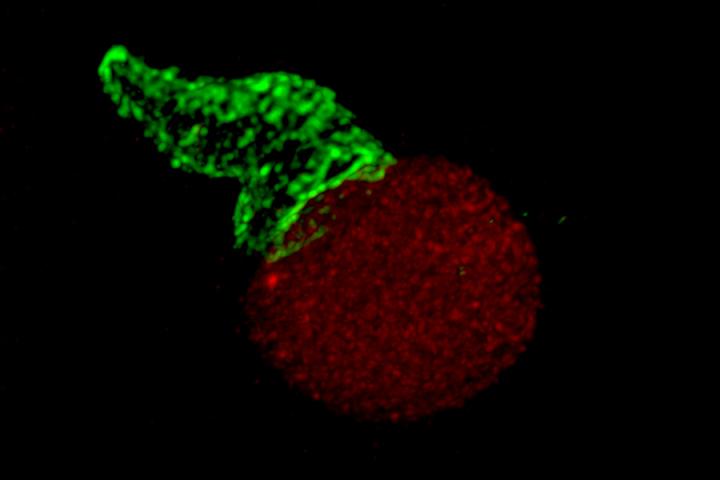
Credit: Photo courtesy of Huang Lab
From testing drugs to developing vaccines, the close study of the immune system is key to improving real-world health outcomes. T-cells are integral to this research, as these white blood cells help tailor the body’s immune response to specific pathogens.
With lattice light-sheet microscopy (LLSM), scientists have been able to closely examine individual cells, such as T-cells, in 4D. But with limited data points, there wasn’t an effective way to analyze the LLSM data.
A new paper by researchers from the Pritzker School of Molecular Engineering (PME) at the University of Chicago, published May 20 in the journal Cell Systems, introduces a solution–a pipeline for lattice light-sheet microscopy multi-dimensional analyses (LaMDA).
The Pritzker Molecular Engineering researchers who authored the paper include graduate students Jillian Rosenberg and Guoshuai Cao of the Huang lab group. They first set out to study T-cell function using high-dimensional microscopy, and then they identified the need for an effective method of analysis. So, they pivoted to developing the LaMDA pipeline.
“We realized that the images are amazing, but they were being underutilized due to a lack of available analysis techniques,” said Cao.
To fill that gap, the researchers decided to shift the paradigm. Instead of treating each cell as a data point, they found a way to treat each molecule as a data point, increasing the total number of data points. This made it possible to conduct a sophisticated analysis.
“We have developed a pipeline to enable machine learning and complex analyses on these videos, which was not previously feasible,” Rosenberg explained. “These analyses allow us to identify differences between molecules that we cannot identify by eye.”
LaMDA could help develop new vaccines
Combining high-dimensional imaging and big data analyses, LaMDA can reveal and even predicT-cellular states. According to Rosenberg, one of the most promising aspects of LaMDA is its potential to predict biological responses, without the need for complex experiments.
More research is needed to further confirm this predictive capacity. However, she explained, “the potential to predicT-cellular states and subcellular signaling is a very powerful asset of LaMDA.”
This means LaMDA could have numerous medical applications, such as drug testing and vaccine development, in addition to expanding the knowledge of T-cell biology.
“Researchers or pharmaceutical companies could use LaMDA to determine how certain drugs are resulting in subtle changes in subcellular signaling, which provides information on both drug safety and efficacy,” said Rosenberg.
“Our LaMDA pipeline could also be extended to the development of peptide vaccines to treat infection, cancer, and autoimmunity,” she added, “or be used to study thymic education or peripheral tolerance, two very important topics in T-cell biology.”
A research tool for T-cells and beyond
Although the researchers began by studying T-cell receptors, creating the LaMDA pipeline quickly became their focus. Using previously known facts about T-cells, they were able to validate LaMDA as an effective analysis pipeline that can be expanded to other fields of study.
“While we ultimately want to learn more about T-cells, our purpose in creating LaMDA was to provide a tool for other researchers to use for future discoveries,” said Rosenberg. The research team intentionally designed LaMDA to be easy for other scientists to use, including those who may be unfamiliar with data science techniques.
“We believe this analysis pipeline will benefit users of high-dimensional microscopy across all fields of science,” said Cao.
It’s worth noting that the researchers have only tested LaMDA in a single molecule, under a few different conditions. “To make this pipeline more robust, it should be validated on other cell types, molecules, and conditions to prove its wide applicability and address any potential unforeseen issues,” Cao said.
One day, the researchers hope the LaMDA pipeline can also be used to study the interaction of multiple molecules.
###
Fernanda Borja-Prieto, an undergraduate student at PME, and Jun Huang, an assistant professor of molecular engineering at the school, are also authors of the paper.
Citation: “Lattice Light-Sheet Microscopy Multi-Dimensional Analyses (LaMDA) of T-Cell Receptor Dynamics Predict T-Cell Signaling States.” Jillian Rosenberg et al. Cell Systems, 2020. DOI: 10.1016/j.cels.2020.04.006
Funding: This work was supported by NIH New Innovator Award 1DP2AI144245 and NSF Career Award 1653782 (To J.H.). J.R. is supported by the NSF Graduate Research Fellowships Program (DGE-1746045).
Media Contact
Ryan Goodwin
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




