Researchers from University of Tsukuba develop a new method that reveals the unique signatures of individual cells in mixtures of bacteria, fungi and yeast under a microscope
Credit: University of Tsukuba
Tsukuba, Japan – When viewed with specialized microscopes, microbial cells show an individual fluorescence pattern, or signature, that depends on the mixture of biomolecules contained within the cells. That complex mixture, with its telltale signature, in turn depends on the type of microbe and its physiological state, such as whether it is growing or what it is consuming as a food source.
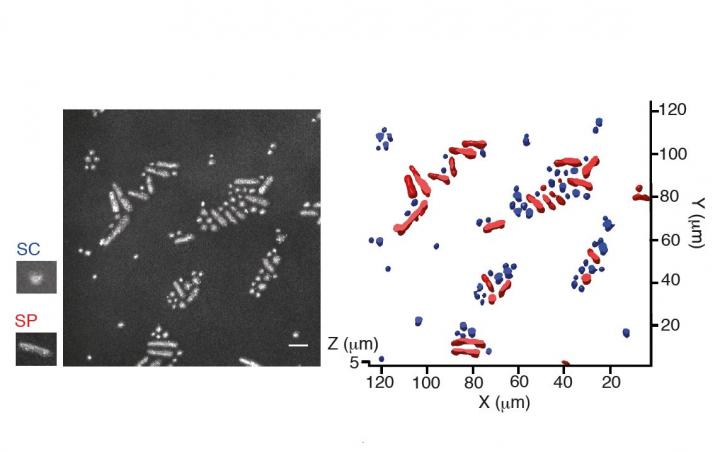
University of Tsukuba researchers have developed a new method, named CRIF (Confocal Reflection microscopy-assisted single-cell Innate Fluorescence), to detect the fluorescence signatures of individual microbial cells. The method is non-destructive, meaning the cells remain alive, and allows cells to be studied in realistic three-dimensional environments. Importantly, the new method can be used to view individual cells within mixtures of different types of microbe, unlike many standard techniques that work best with “pure” populations where the cells are all alike. The team recently published their findings in Applied and Environmental Microbiology.
“We used a confocal microscope, which can generate images from three-dimensional materials–rather like comparing a sequence of slices to build up an image of a whole intact structure. This enabled us to find the locations of the microbial cells within the sample,” says lead and co-corresponding author Yutaka Yawata. “Combining this with spectroscopy to measure a set of fluorescence signals under the microscope, we were able to see the different types of microbes within the sample, and read their signatures to identify the individual cells.”
The team went on to train computer models to read the fluorescence signatures and distinguish different types of cells. The models learned to recognize cells automatically, even when looking at different cells with very similar shapes and sizes, and analyze the images for cell signatures to identify the cells according to their type and physiological state.
“Our technique to recognize and track the innate fluorescence signatures of each of the individual cells within three-dimensional samples opens up exciting new opportunities to explore mixed microbial populations, as found in natural environments,” says co-corresponding author Nobuhiko Nomura. “It will help researchers understand how microbes grow and interact with each other in the real world.”
###
Media Contact
Naoko Yamashina
[email protected]
Related Journal Article
http://dx.




