In a groundbreaking exploration into the metabolic underpinnings of clear cell renal cell carcinoma (ccRCC), researchers have unveiled a detailed landscape of altered biochemical pathways that could transform the way oncologists predict and monitor therapeutic responses. This study, recently published in BMC Cancer, provides a sophisticated metabolic classification for ccRCC based on extensive profiling of tumor and adjacent normal tissue samples, offering fresh avenues for biomarker discovery and personalized medicine in cancer treatment.
Clear cell renal cell carcinoma, noted for its frequent biallelic inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene, has long been a subject of intense study due to its complex metabolic rewiring and resistance to standard therapies. The loss of VHL function disrupts key regulatory circuits, significantly influencing cellular metabolism and substrate utilization. In this comprehensive study, the authors embarked on validating previously identified metabolomic signatures and teasing out metabolic predictors that correlate with patient responses to systemic treatments such as VEGF-tyrosine kinase inhibitors (VEGF-TKI) and immune checkpoint blockade (ICB).
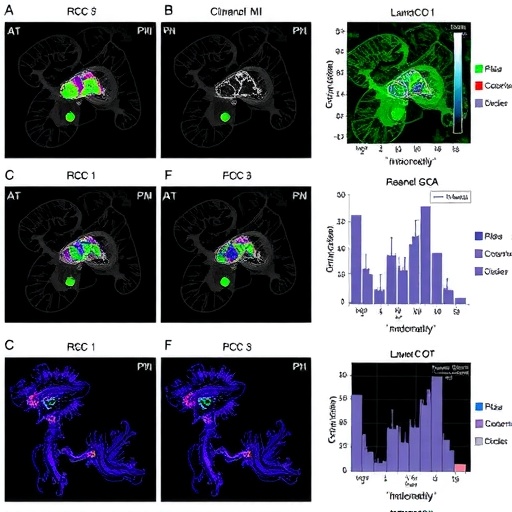
The research team meticulously analyzed 52 paired tumor and normal kidney samples utilizing advanced metabolomic profiling techniques. This paired design allowed a controlled comparison, ensuring that intrinsic patient variability did not confound the differential metabolic landscape observed in ccRCC tissues. Using paired t-tests and unsupervised clustering algorithms, the researchers stratified tumors into four distinct metabolic subgroups, each characterized by unique metabolites and pathways.
.adsslot_QB40GJX1tL{ width:728px !important; height:90px !important; }
@media (max-width:1199px) { .adsslot_QB40GJX1tL{ width:468px !important; height:60px !important; } }
@media (max-width:767px) { .adsslot_QB40GJX1tL{ width:320px !important; height:50px !important; } }
ADVERTISEMENT
One of the salient findings was the consistent activation of the upper glycolytic pathway and the pentose phosphate pathway (PPP) across tumor samples. These metabolic circuits are essential for providing cancer cells with the biosynthetic precursors and reducing equivalents necessary for rapid proliferation and survival under oxidative stress. The elevated glutamine levels detected further reinforce the notion that ccRCC cells shift towards glutamine addiction, fueling both anaplerosis and redox balance. Intriguingly, proteinogenic amino acids, other than glutamine, were found to be diminished, hinting at a selective metabolic remodeling that privileges certain substrates over others.
Despite prior reports emphasizing lactate accumulation as a hallmark of ccRCC metabolism, this investigation revealed a pronounced heterogeneity in lactate concentrations across the metabolic subgroups. This variability suggests that lactate production and clearance may be more nuanced than previously appreciated, potentially reflecting adaptation to microenvironmental conditions or divergent metabolic dependencies among tumor cells.
Further linking metabolism with pathophysiology, the metabolic clusters enriched with high-grade tumors exhibited decreased expression of vascular endothelial growth factor (VEGF) pathway-related genes. This observation is clinically relevant, as VEGF signaling is a pivotal mediator of angiogenesis in ccRCC, and its downregulation could impact both tumor aggressiveness and therapeutic targets.
Diving deeper into therapy-specific metabolomic alterations, the study analyzed specimens from patients treated with VEGF-TKI and immune checkpoint inhibitors separately. VEGF-TKI responders displayed distinctive decreases in certain fatty acid species, aligning with previous evidence that fatty acid metabolism may modulate angiogenic signaling and drug sensitivity. Conversely, patients responding to immune checkpoint blockade exhibited a unique metabolic fingerprint marked by depleted tryptophan and hydroquinone levels, alongside increases in metabolites such as pyruvic acid-oxime, 3-hydroxypropinoic acid, and hydroxylamine. These changes are particularly intriguing given the immunometabolic crosstalk underpinning anti-tumor immunity and the role of tryptophan metabolism in immune evasion.
The implications of these findings are manifold. By validating a robust metabolomic classification of ccRCC, this study not only augments our understanding of tumor biology but also underlines the utility of metabolic biomarkers for predicting patient response to diverse systemic therapies. Such biomarkers could be incorporated into clinical workflows to tailor treatments, optimize drug selection, and monitor efficacy non-invasively.
Methodologically, the integration of metabolomics with transcriptomic data and clinical parameters exemplifies a systems biology approach crucial for unraveling cancer heterogeneity. The four metabolic subgroups defined could serve as a foundation for future trials aimed at stratifying patients based on metabolic vulnerabilities, enhancing precision oncology strategies.
Moreover, the identification of differential fatty acid and amino acid metabolism in relation to therapeutic outcomes opens exciting prospects for metabolic reprogramming interventions. Targeting aberrant glutamine metabolism, modulating lactate production, or correcting amino acid imbalances might enhance the efficacy of existing treatments or overcome resistance mechanisms.
From a broader perspective, this research illustrates the increasingly recognized role of the tumor microenvironment and metabolic plasticity in cancer progression. ccRCC’s metabolic landscape is shaped not only by genetic mutations but also by the adaptive responses to hypoxia and nutrient availability, encapsulated by the VHL-driven changes elucidated here.
One cannot ignore the translational potential of these insights. As systemic therapies expand with novel agents entering the clinic, the ability to predict which patients will benefit most from VEGF-TKI or ICB regimens based on metabolomic signatures could revolutionize care paradigms, reduce unnecessary toxicity, and improve survival outcomes.
Furthermore, the study invites a reevaluation of lactate’s role as a universal biomarker in ccRCC. Given the diverse lactate levels observed, future investigations should explore the mechanisms governing lactate metabolism’s heterogeneity and its link to immune infiltration and stromal interactions.
In conclusion, this meticulous dissection of ccRCC’s metabolic landscape underscores how cancer cells orchestrate complex biochemical adaptations to flourish and evade therapy. The discovery of metabolite markers associated with drug response heralds a new era of metabolomics-driven oncology, wherein small molecules within the tumor milieu could become pivotal guides for therapeutic decision-making. It is an exciting time for cancer research, with metabolism emerging from the shadows to take center stage in the quest for more effective and personalized treatments.
Subject of Research: Metabolic profiling and classification of clear cell renal cell carcinoma with identification of metabolites predictive of response to systemic therapies.
Article Title: Metabolic landscape of clear cell renal cell carcinoma and search for metabolites predictive of drug response.
Article References:
Ozawa, M., Naito, S., Makinoshima, H. et al. Metabolic landscape of clear cell renal cell carcinoma and search for metabolites predictive of drug response.
BMC Cancer 25, 1357 (2025). https://doi.org/10.1186/s12885-025-14661-4
Image Credits: Scienmag.com
DOI: https://doi.org/10.1186/s12885-025-14661-4
Keywords: Clear cell renal cell carcinoma, ccRCC, metabolomics, metabolic biomarkers, VHL gene, VEGF-TKI, immune checkpoint blockade, tumor metabolism, glycolysis, pentose phosphate pathway, glutamine metabolism, fatty acids, tryptophan metabolism, personalized oncology
Tags: advanced metabolomic techniquesbiochemical pathways in ccRCCbiomarkers for personalized medicineclear cell renal cell carcinomaimmune checkpoint blockade responsemetabolic profiling in cancermetabolomic signatures in oncologyrenal cancer treatment advancementstherapeutic response predictiontumor metabolism alterationsVEGF-tyrosine kinase inhibitorsVHL tumor suppressor gene