Gladstone researchers used a machine-learning approach to discover new ways of controlling the spatial organization of induced pluripotent stem cells
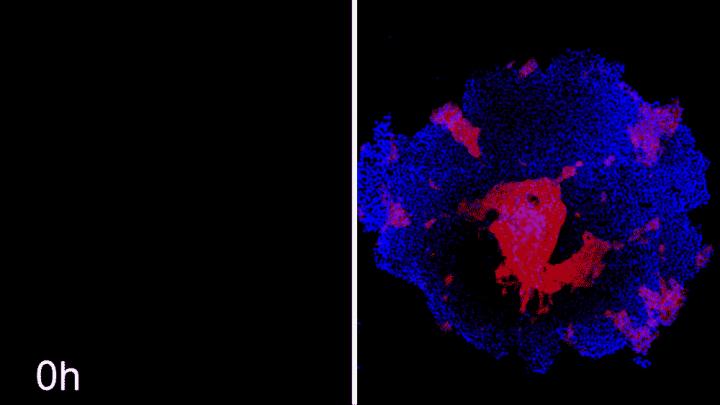
Credit: Photo credit: Ashley Libby, David Joy, and Iman Haghighi, Gladstone Institutes
SAN FRANCISCO, CA November 20, 2019– Model organs grown from patients’ own cells may one day revolutionize how diseases are treated. A person’s cells, coaxed into heart, lung, liver, or kidney in the lab, could be used to better understand their disease or test whether drugs are likely to help them. But this future relies on scientists’ ability to form complex tissues from stem cells, a challenging undertaking.
In their natural environment, stem cells form predictable patterns as they mature; over time, these patterns morph into the tissues of an adult organism. In the lab though, researchers have struggled to control the spatial organization of stem cells–an important step toward being able to create functional organs for research or therapeutic purposes. Some have turned to 3-D printing to lay out populations of stem cells in a desired shape. But the approach isn’t always successful, with cells often migrating away from their printed locations.
Now, scientists at Gladstone Institutes, in collaboration with researchers at Boston University, have used a computational model to learn how to coax stem cells into forming new arrangements, including those that might eventually be useful in generating personalized organs.
“We’ve shown how we can leverage the intrinsic ability of stem cells to organize,” said Gladstone Senior Investigator Todd McDevitt, PhD, a lead author of the study, which was published in the journal Cell Systems. “This gives us a new way of engineering tissues, rather than a printing approach where you try to physically force cells into a specific configuration.”
“This works exemplifies the power of applying a computational approach to stem cell biology to make sense of the complexity in these cells,” said Calin Belta, director of the Boston University Robotics Lab and co-corresponding author on the new paper.
Induced pluripotent stem (iPS) cells, similar to the stem cells found in an embryo, have the potential to become nearly every type of cell in the body. Researchers have found ways to direct iPS cells to become many of these cell types, including heart and brain. Some are already using these cells to model diseases in the lab or even transplant into patients. But clumps of cells in a Petri dish aren’t the same thing as functioning three-dimensional organs.
“Despite the importance of organization for functioning tissues, we as scientists have had difficulty creating tissues in a dish with stem cells,” said Ashley Libby, a graduate student in the UC San Francisco Developmental & Stem Cell Biology Program and co-first author of the new paper, who worked on the project with David Joy, a graduate student in the joint Graduate Program in Bioengineering from UC Berkeley and UC San Francisco (BioE). “Instead of an organized tissue, we often get a disorganized mix of different cell types.”
McDevitt and his colleagues previously showed that blocking, or “knocking down,” the expression of two different genes, ROCK1 and CDH1, affected the layout of iPS cells grown in a Petri dish. The scientists wondered whether they could predict the exact arrangement of cells that would result from altering ROCK1 and CDH1 by different degrees at different timepoints. But there were so many possible variables–the timing and degree of each gene knockdown, the duration of the experiment, the proportion of cells affected–that testing every possible combination would be too time-consuming. So McDevitt’s group teamed up with the Belta Lab who could help them create a model to do the job.
The researchers used a CRISPR/Cas9 gene-editing system that could be induced to block expression of ROCK1 or CDH1 at any time during an experiment by adding a drug to the iPS cells. In addition, they engineered the system so that cells fluoresced in different colors when they lost expression of ROCK1 or CDH1, making it easier to study changes to the arrangement of the cells.
McDevitt’s group carried out a handful of experiments using different doses and timing of the CRISPR/Cas9 system. Then, the computational researchers started inputting the results into a machine-learning program, designed to identify patterns within a dataset.
“Machine learning can predict what movie you might like based on your viewing history, but it can also generate new insights into biological systems by mimicking them.” said Demarcus Briers, co-first author of the new paper who performed the work during his graduate studies at Boston University. “Our machine-learning model allows us to predict new ways that stem cells can organize themselves, and produces instructions for how to recreate these predictions in the lab.”
The machine-learning program used results from the initial stem cell experiments to infer ways that ROCK1 and CDH1 affect iPS cell organization. With the model up and running, the researchers then began to probe whether it could compute how to make entirely new patterns, like a bull’s-eye or an island of cells.
“The power of this model is that it can generate thousands of data points simulating things that it could take months for me to do in a lab,” said Libby.
The simulations narrowed down a set of starting conditions that might lead to the desired arrangement of cells–informing researchers exactly when, where, and how to add drugs to their iPS cells to shut off ROCK1 and CHD1. Then, McDevitt and Libby could test those suggested conditions. The machine-learning system, it turned out, was correct–at least when it came to the bull’s-eye and island patterns they were after. In the lab, for the first time, the researchers were able to reliably generate concentric circles of stem cell populations looped around each other.
“I was just blown away when I first saw the results,” said Bruce Conklin, MD, a Gladstone senior investigator who also worked on the new study. “Modeling cell behavior is the Holy Grail of biology and this paper takes an important step forward in doing that.”
The team would like to expand the model in the future–adding in the effects of other developmental genes to get an even wider variety of possible cell configurations. They also plan to work toward designing three-dimensional shapes in addition to the two-dimensional layouts they’ve already studied.
“We’re now on the path to truly engineering multicellular organization, which is the precursor to engineering organs,” said McDevitt, who is also the director of the BioE graduate program. “When we can create human organs in the lab, we can use them to study aspects of biology and disease that we wouldn’t otherwise be able to.”
###
About the Study
The paper, “Self-Organized Pluripotent Stem Cell Patterning by Automated Design” will be published by the journal Cell Systems on November 20, 2019.
Other authors include: Iman Haghighi from Boston University.
The work at Gladstone was supported by the National Science Foundation, the California Institute of Regenerative Medicine, and the National Heart, Lung, and Blood Institute.
Media Contact
Megan McDevitt
[email protected]
415-734-2019
Related Journal Article
http://dx.




