Brain tissue is one of the most intricate specimens that scientists have arguably ever dealt with. Packed with currently immeasurable amount of information, the human brain is the most sophisticated computational device with its network of around 86 billion neurons. Understanding such complexity is a difficult task, and hence making progress requires technologies to unravel the tiny, complex interactions taking place in the brain at microscopic scales. Imaging is therefore an enabling tool in neuroscience.
Credit: © Johann Danzl
Brain tissue is one of the most intricate specimens that scientists have arguably ever dealt with. Packed with currently immeasurable amount of information, the human brain is the most sophisticated computational device with its network of around 86 billion neurons. Understanding such complexity is a difficult task, and hence making progress requires technologies to unravel the tiny, complex interactions taking place in the brain at microscopic scales. Imaging is therefore an enabling tool in neuroscience.
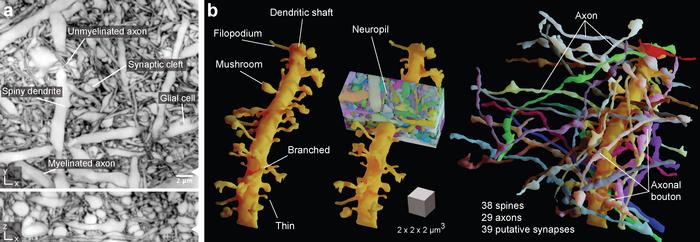
The new imaging and virtual reconstruction technology developed by Johann Danzl’s group at ISTA is a big leap in imaging brain activity and is aptly named LIONESS – Live Information Optimized Nanoscopy Enabling Saturated Segmentation. LIONESS is a pipeline to image, reconstruct, and analyze live brain tissue with a comprehensiveness and spatial resolution not possible until now.
“With LIONESS, for the first time, it is possible to get a comprehensive, dense reconstruction of living brain tissue. By imaging the tissue multiple times, LIONESS allows us to observe and measure the dynamic cellular biology in the brain take its course,” says first author Philipp Velicky. “The output is a reconstructed image of the cellular arrangements in three dimensions, with time making up the fourth dimension, as the sample can be imaged over minutes, hours, or days,” he adds.
and achieve high-resolution 3D imagery
without damaging the living sample.
Collaboration and AI the Key
The strength of LIONESS lies in refined optics and in the two levels of deep learning – a method of Artificial Intelligence – that make up its core: the first enhances the image quality and the second identifies the different cellular structures in the dense neuronal environment.
The pipeline is a result of a collaboration between the Danzl group, Bickel group, Jonas group, Novarino group, and ISTA’s Scientific Service Units, as well as other international collaborators. “Our approach was to assemble a dynamic group of scientists with unique combined expertise across disciplinary boundaries, who work together to close a technology gap in the analysis of brain tissue,” Johann Danzl of ISTA says.
Surpassing hurdles
Previously it was possible to get reconstructions of brain tissue by using Electron Microscopy. This method images the sample based on its interactions with electrons. Despite its ability to capture images at a few nanometers—a millionth of a millimeter—resolution, Electron Microscopy requires a sample to be fixed in one biological state, which needs to be physically sectioned to obtain 3D information. Hence, no dynamic information can be obtained.
Another previously known technique of Light Microscopy allows observation of living systems and record intact tissue volumes by slicing them “optically” rather than physically. However, Light Microscopy is severely hampered in its resolving power by the very properties of the light waves it uses to generate an image. Its best-case resolution is a few hundred nanometers, much too coarse-grained to capture important cellular details in brain tissue.
Using Super-resolution Light Microscopy scientists can break this resolution barrier. Recent work in this field, dubbed SUSHI (Super-resolution Shadow Imaging), showed that applying dye molecules to the spaces around cells and applying the Nobel Prize-winning super-resolution technique STED (Stimulated Emission Depletion) microscopy reveals super-resolved ‘shadows’ of all the cellular structures and thus visualizes them in the tissue. Nevertheless, it has been impossible to image entire volumes of brain tissue with resolution enhancement that matches the brain tissue’s complex 3D architecture. This is because increasing resolution also entails a high load of imaging light on the sample, which may damage or ‘fry’ the subtle, living tissue.
Herein lies the prowess of LIONESS, having been developed for, according to the authors, “fast and mild” imaging conditions, thus keeping the sample alive. The technique does so while providing isotropic super-resolution—meaning that it is equally good in all three spatial dimensions—that allows visualization of the tissue’s cellular components in 3D nanoscale resolved detail.
LIONESS collects only as little information from the sample as needed during the imaging step. This is followed by the first deep learning step to fill in additional information on the brain tissue’s structure in a process called Image Restoration. In this innovative way, it achieves a resolution of around 130 nanometers, while being gentle enough for imaging of living brain tissue in real-time. Together, these steps allow for a second step of deep learning, this time to make sense of the extremely complex imaging data and identify the neuronal structures in an automated manner.
Homing In
“The interdisciplinary approach allowed us to break the intertwined limitations in resolving power and light exposure to the living system, to make sense of the complex 3D data, and to couple the tissue’s cellular architecture with molecular and functional measurements,” says Danzl.
For virtual reconstruction, Danzl and Velicky teamed up with visual computing experts: the Bickel group at ISTA and the group led by Hanspeter Pfister at Harvard University, who contributed their expertise in automated segmentation—the process of automatically recognizing the cellular structures in the tissue—and visualization, with further support by ISTA’s image analysis staff scientist Christoph Sommer. For sophisticated labeling strategies, neuroscientists and chemists from Edinburgh, Berlin, and ISTA contributed. Consequently, it was possible to bridge functional measurements, i.e. to read out the cellular structures together with biological signaling activity in the same living neuronal circuit. This was done by imaging Calcium ion fluxes into cells and measuring the cellular electrical activity in collaboration with the Jonas group at ISTA. The Novarino group contributed human cerebral organoids, often nicknamed mini-brains that mimic human brain development. The authors underline that all of this was facilitated through expert support by ISTA’s top-notch scientific service units.
Brain structure and activity are highly dynamic; its structures evolve as the brain performs and learns new tasks. This aspect of the brain is often referred to as “plasticity”. Hence, observing the changes in the brain’s tissue architecture is essential to unlocking the secrets behind its plasticity. The new tool developed at ISTA shows potential for understanding the functional architecture of brain tissue and potentially other organs by revealing the subcellular structures and capturing how these might change over time.
Journal
Nature Methods
DOI
10.1038/s41592-023-01936-6
Method of Research
Imaging analysis
Subject of Research
Human tissue samples
Article Title
Dense 4D nanoscale reconstruction of living brain tissue
Article Publication Date
10-Jul-2023



