Credit: (Picture: Team Markus Sauer / University of Wuerzburg)
With high-resolution microscopy, it is theoretically possible to image cell structures with a resolution of a few nanometres. However, this has not yet been possible in practice.
The reason for this is that antibodies carrying a fluorescent dye are usually used to label cell structures. Therefore, the dye is not located directly at the target structure, but about 17.5 nanometres away from it. Partly because of this distance error, the theoretically achievable resolution could not be achieved so far.
Publication in Nature Communications
An international research team has now overcome this hurdle. This was achieved by combining the super-resolution microscopy methods dSTORM and expansion microscopy (ExM). The journal Nature Communications presents the results.
The publication was led by a team from the Biocenter of Julius-Maximilians-Universität (JMU) Würzburg in Bavaria, Germany: Professor Markus Sauer, Head of the Department of Biotechnology and Biophysics, with PhD students Fabian Zwettler and Sebastian Reinhard. Professors Paul Guichard from the University of Geneva (Switzerland) and Toby Bell from Monash University (Australia) also played a key role.
Obstacles to combining dSTORM and ExM
The dSTORM method, developed in Professor Sauer’s group, achieves an almost molecular resolution of about 20 nanometers. To further increase the resolution, a combination with expansion microscopy, which has been available for a few years now, seemed promising.
In ExM, the sample to be examined is cross-linked into a swellable polymer. Then the interactions of the molecules in the sample are destroyed and the sample is allowed to swell in water. This leads to an expansion: the molecules to be imaged drift spatially apart by a factor of four.
Why the two methods could not be combined until now:
- The fluorescent dyes used for dSTORM to label the molecules did not survive the polymerization of the aqueous gel.
- A buffer solution is needed for dSTORM, but the expanded sample shrinks to its original size in such buffers.
Distance error significantly reduced
“By stabilizing the gel and immune staining only after expansion, we could overcome these hurdles and successfully combine the two microscopy methods,” says Markus Sauer. As a result, the distance error melts to just five nanometers when expanded 3.2 times. This makes fluorescence imaging with molecular resolution possible for the first time.
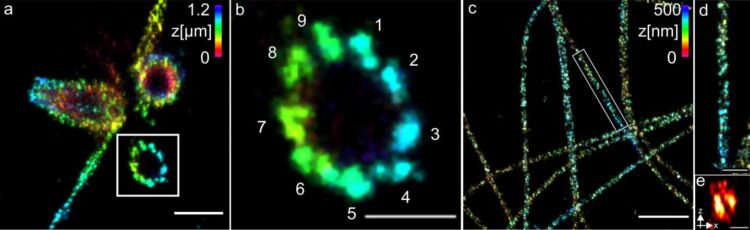
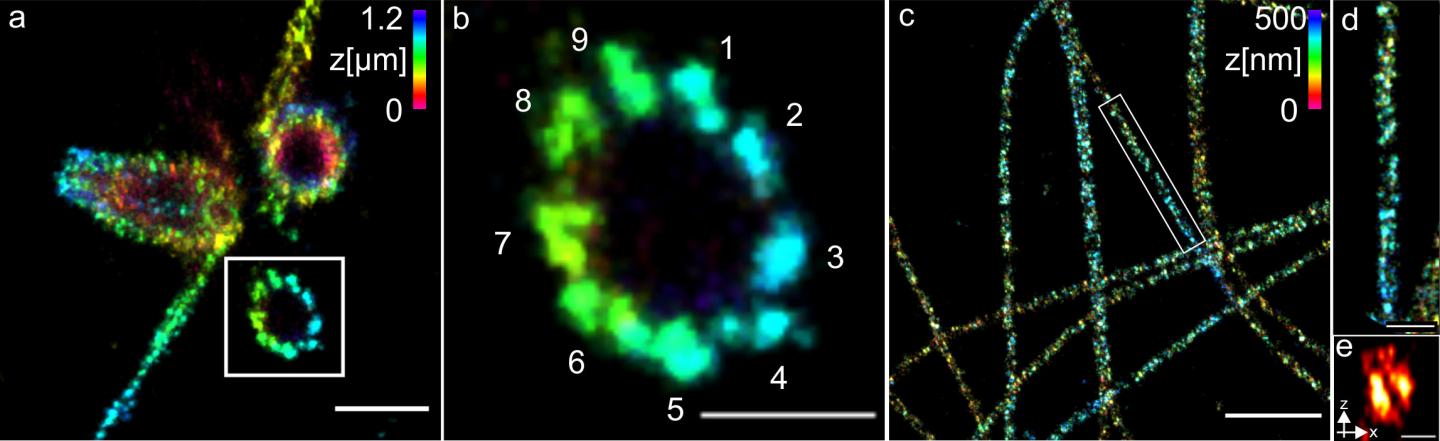
The researchers used centrioles and structures that are composed of the protein tubulin to show how well their method works. They were able to visualise tubulin tubes as hollow cylinders with a diameter of 25 nanometres. The researchers succeeded in sharply imaging groups of three made up of tubulin structures at a distance of 15 to 20 nanometres at the centrioles. The centrioles are cell structures that play an important role in cell division.
Professor Sauer’s conclusion: “For many important cell components, the combination of ExM and dSTORM now enables us to gain detailed insights into molecular function and architecture for the first time. The team therefore plans to apply the method to different structures, organelles and multiprotein complexes of the cell.
###
Media Contact
Prof. Dr. Markus Sauer
[email protected]
Original Source
https:/
Related Journal Article
http://dx.