Flipon-based Computers
Credit: Alan Herbert
Dr. Herbert asks what if our genome was far smarter than everyone previously believed? What if in the many DNA repeat elements lay the foundation for building a novel type of biocomputer? This approach would enable calculations performed with self-renewing wetware rather than stone-cold hardware, opening the door to logic circuits based on DNA that flips from one state to another, analogous to the way silicon switches “on” and “off”.
How might this kind of DNA computer work? Simple repeats are called that because the DNA sequence repeats itself over and over again a number of times. The repeats actually are great for building DNA structures with useful properties. Instead of the everyday B-DNA described by Watson and Crick, repeats of certain DNA sequences can morph into some rather exotic higher-energy 3D structures. They can form left-handed duplexes, three-stranded triplexes, and four-stranded quadruplexes. The different DNA structures change how information is read out from DNA to make RNA.
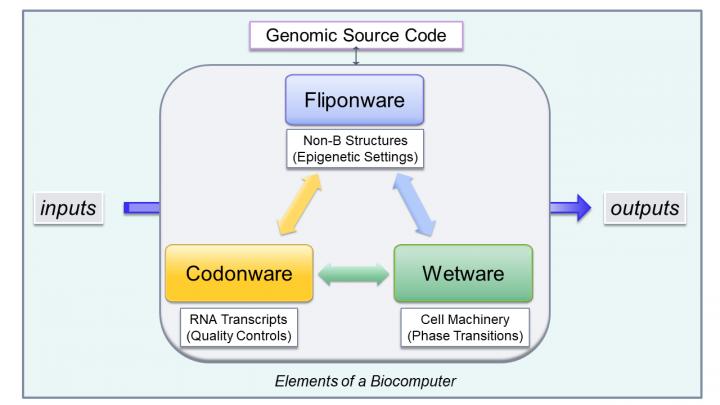
Those repeat sequences that are able to switch from one DNA structure to another are called flipons. Collectively, Dr. Herbert refer to these flipons and their associated functionality as fliponware. Fliponware sets the stage for a cell to make the right wetware tools and run the right genetic program to get the job at hand done. It allows assembly of gene pieces into different RNA-based programs for a cell to overcome challenges from its environment.
Dr. Herbert provides examples of how flipons can be used to create genetic programs, describing ways they can be wired into the logic gates that computers need to function. The flipon logic gates can perform ‘AND’ or ‘NOT’ operations. Many can be combined to perform the Boolean operations essential to a universal computer as first described by Turing. Their use in the genome for logical operations resembles how Turing machines work, but instead of a Turing tape, the cell uses RNA to record the results. The series of processing steps decides whether the RNA message is stable or not and the genetic program to execute.
Many of the novel RNA assemblies are trashed without ever being used–they may not be needed or their assembly is defective (equivalent to a FALSE result from a Turing machine). Those messages that persist become the codonware of the cell (logically the same as a TRUE result from a Turing machine). The codonware that results then defines the cell’s biology by the wetware they direct the production of..
This form of computation differs from previously described DNA computers built without flipons. These other devices depend on whether or not pairs of DNA sequences match each other. If they do, the matches enable the next calculation step. Flipon computers differ in many ways from those using DNA matching. First, some flipons switch very fast (in milliseconds) because all they need to do is change from one 3D structure to another without the need to search for a matching DNA to pair with. Turning a flipon ON or OFF is possible in many different ways. For example, just stretching the DNA can cause flipons to morph, or they can flip due to a change in temperature or variations in salt concentration.
The combination of fliponware, wetware and codonware is analogous to program code, machine language and hardware in a silicon computer. Each of these bioware sets has memory to enable learning. They use error correction to prevent bad outcomes. While flipons are made with simple sequences, codonware is made from complex sequence mixes. The information provides the cell very detailed information on the wetware to make. In simple terms, fliponware instructs which tools to use for a particular job, codonware tells how to make the tools, and wetware does the job.
Dr. Herbert adds, “I expect that soon there will be many exciting and beneficial applications for fliponware. Simple sequences are not your grandparent’s junk–instead they are like the rules of thumb that simplify life, acquired over many years of evolution.”
Fliponware has several immediate applications:
- 1. Therapeutic applications (target drugs to flipon states that enable cancers or inflammatory diseases)
2. Biosensors (to detect environmental changes)
3. Persistent DNA memory (exploiting the extreme stability of quadruplexes after they form)
4. Cellular switches (change of output in response to a change of input)
5. Novel DNA nano-architectures (the 3D structure formed depends on flipon state)
###
About InsideOutBio
InsideOutBio is a start-up focused on developing next generation of cancer therapeutics to ‘light’ up tumors for the immune system to kill by reprogramming self/nonself pathways within cancer cells. Dr. Herbert leads discovery at InsideOutBio. His work on Z-DNA was foundational to the discovery of flipons. These statements about InsideOutBio comply with Safe-Harbor laws. They are forward-looking and involve known and unknown risks and uncertainties. They are not guarantees of future performance and undue reliance should not be placed on them.
Media Contact
Alan Herbert
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




