Evolutionary biology: Publication in Communications Biology
Credit: HHU / Joana Xavier
Roughly five years ago, Institute Head Prof. Dr. William (Bill) Martin and his team introduced the last universal common ancestor of all living organisms and named it “LUCA”. It lived approximately 3.8 billion years ago in hot deep sea hydrothermal vents.
Now the evolutionary biologists in Duesseldorf have described a further ancient cell named “LBCA” (“Last Bacterial Common Ancestor”). It is the ancestor of today’s largest domain of all living organisms: Bacteria. In Communications Biology, they report on their new research approaches which led to the successful prediction of the biochemistry of LBCA and its phylogenetic links.
Bacteria are almost as old as life itself. LBCA lived around 3.5 billion years ago in a similar environment to LUCA. In order to unlock LBCA’s genetic code, its properties and its story, the research team examined the genomes of 1,089 bacterial anaerobes or bacteria that survive without oxygen. “Abandoning aerobes made sense for our work”, explains first author Dr. Joana C. Xavier. “If bacteria originated at a time when the Earth was anoxic, it does not make sense to investigate their origin considering species full of adaptations caused by oxygen.”
Higher life forms pass on their genetic code from parent to offspring via vertical gene transfer. As a result, the genome provides information on phylogenetic history. But bacteria are masters in another form of gene transfer, namely lateral gene transfer (LGT). This allows bacteria to exchange genetic information across different strains. This posed a major challenge in reconstructing the LBCA genome, as it renders the traditional phylogenetic methods incapable of inferring the root in the bacterial evolutionary tree.
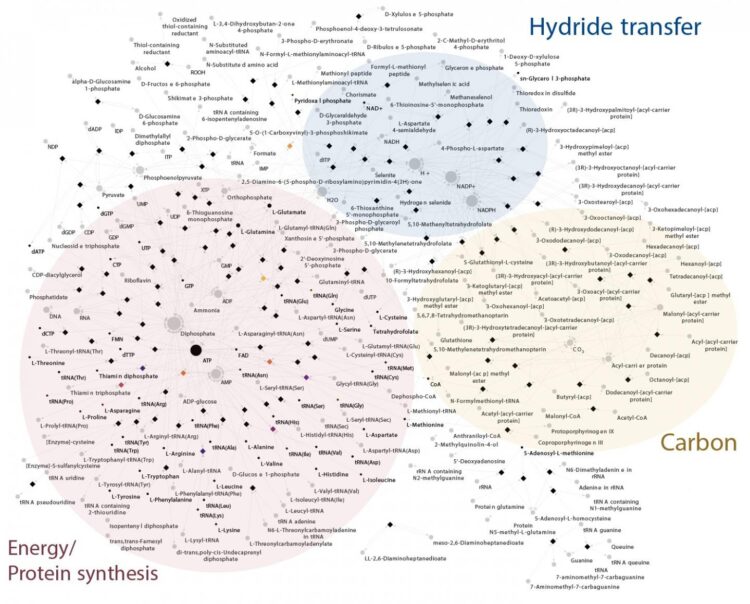
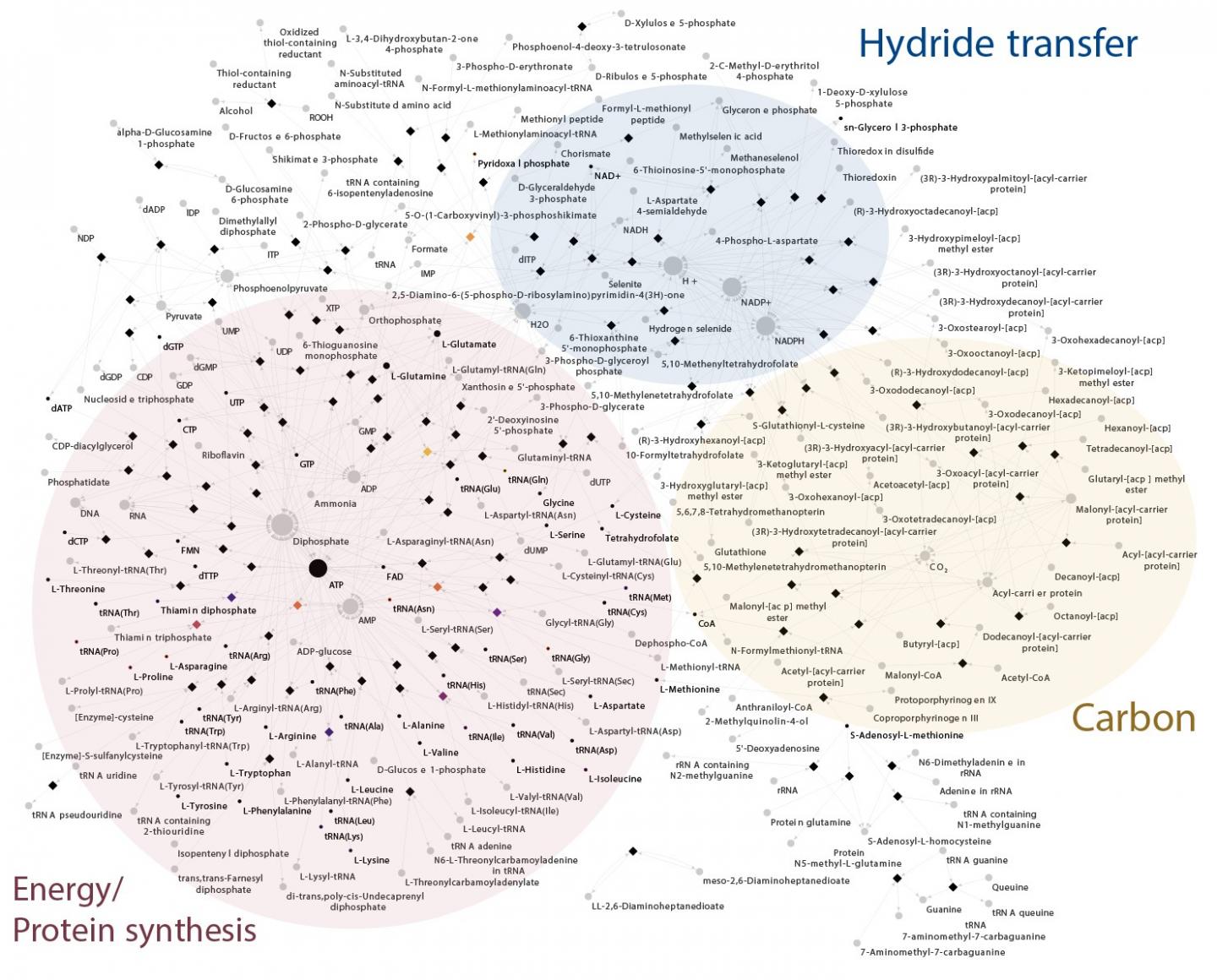
For this reason, the researchers in Duesseldorf used biochemical networks together with thousands of individual trees. They investigated 1,089 anaerobic genomes and identified 146 protein families conserved in all bacteria. These proteins make up a nearly complete core metabolic network.
To complete LBCA’s biochemistry, just nine further genes had to be added for the reconstructed metabolic network to include all essential and universal metabolites. To be fully independent and self-generated, LBCA’s network would still require further genes inherited from the last universal common ancestor, LUCA, and nutrients from the environment.
With LBCA’s metabolic network in hand, the authors then used statistical methods to determine which of the modern bacterial groups are most similar to LBCA. They did this using a method called Minimal Ancestor Deviation, MAD, previously developed by one of the co-authors, Fernando D. K. Tria: “The analyses revealed that the earliest branch of Bacteria to diverge was most similar to modern Clostridia, followed closely by Deltaproteobacteria, Actinobacteria and some members of Aquifex. In common, these groups have the acetyl-CoA pathway for carbon fixation and/or energy metabolism.”
Prof. William Martin, senior author of the study, explains: “This is the only carbon fixation pathway present in both archaea and bacteria and that traces to LUCA. This result, obtained independently, is also in line with our most recent findings on the origin and early evolution of life in hydrothermal vents.”
“We can infer with confidence that LBCA was most likely rod-shaped”, says Xavier. “If it was similar to Clostridia, it is possible that LBCA was able to sporulate.” This hypothesis was recently laid out by other researchers “and is highly compatible with our results”, says Xavier. Forming spores would allow early cells to survive the inhospitable environment of the early Earth.
###
The authors went further to determine if lateral gene transfer played a role in the era of LBCA. Fernando D. K. Tria explains: “LBCA genes are much more vertical than your average bacterial gene, which indicates that they are very ancient.”
Original publication
Xavier, J.C., Gerhards, R.E., Wimmer, J.L.E. et al., The metabolic network of the last bacterial common ancestor, Commun Biol 4, 413 (2021).
DOI: 10.1038/s42003-021-01918-4
Media Contact
Dr. Arne Claussen
[email protected]
Original Source
https:/
Related Journal Article
http://dx.