Credit: Tokyo Tech
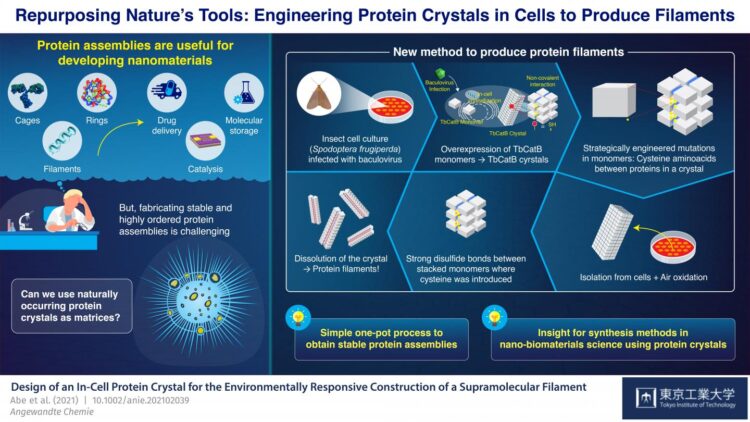
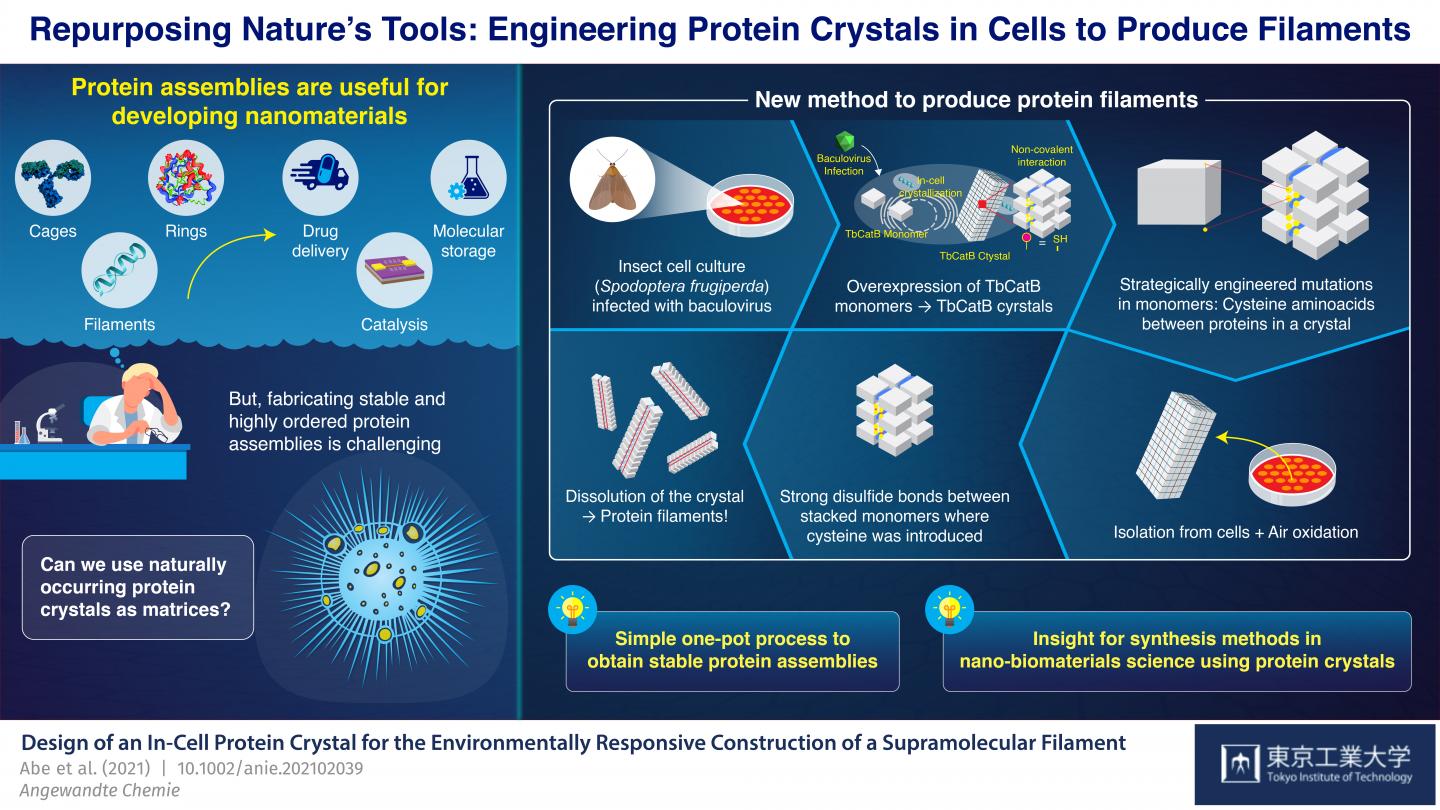
Proteins are undoubtedly some of the most fascinating biomolecules, and they perform many of the functions that (in our eyes) separate life from inanimate matter. Multi-molecular protein assemblies even have large-scale structural functions, as evidenced by feathers, hair, and scales in animals. It should come as no surprise that, with progress in advanced nanotechnology and bioengineering, artificial protein assemblies have found applications in a variety of fields, including catalysis, molecular storage, and drug delivery systems.
However, producing ordered protein assemblies remains challenging. It is particularly difficult to get monomers, the building blocks of proteins, to assemble stably into the desired structures; this generally requires very accurate design and control of synthesis conditions, such as pH (acidity) and temperature. Recent studies found ways to circumvent this problem by using protein crystals–solid molecular arrangements that occur naturally in some organisms–as precursor matrices to produce protein assemblies.
At Tokyo Institute of Technology, Japan, a team of scientists led by Professor Takafumi Ueno has been working on a promising approach for synthesizing protein assemblies from protein crystals. Their strategy involves introducing mutations into the genetic code of an organism that naturally produces protein crystals. These mutations cause disulfide bonds (S-S) to form between monomers in very specific locations in the crystals. The crystals are then dissolved, but instead of breaking down completely into their individual monomers as usual, the newly introduced S-S bonds hold groups of monomers together and the crystals split into many of the desired protein assemblies. With this approach, Ueno’s team has managed to synthesize protein cages and tubes by essentially using living cells as nano-3D printers.
In their latest study, which was published in Angewandte Chemie International Edition, the team demonstrated yet another application of their novel strategy; this time for the synthesis of bundled protein filaments. They used a culture of insect cells (Spodoptera frugiperda) infected with a virus that caused overexpression of a monomer called “TbCatB.” These monomers naturally aggregate inside the cells into protein crystals, which are held together there by the relatively weak non-covalent interactions between monomers. The scientists strategically introduced two mutations in the cells so that each monomer had two thiol groups (-SH) of cysteine at critical interface points with other monomers.
The crystals were extracted from the cells and left to oxidize at room temperature, which caused the thiol groups to change into strong S-S bonds between monomers adjacent along a single direction by autoxidation under air. When the crystals were dissolved, these disulfide bonds, together with some lingering non-covalent interactions, resulted in the formation of bundled protein filaments that were two monomers wide–about 8.3 nanometers. “With our strategy, we achieved a highly precise arrangement of protein molecules while suppressing random aggregation of monomers due to unwanted sulfide bonds, all in a relatively straightforward one-pot process,” highlights Ueno.
Overall, the approach demonstrated by the team at Tokyo Tech stands as an innovative way to synthesize protein structures via rational genetic engineering and by using the tools naturally available to cells of certain organisms. “We consider our synthesis method a useful advance in nano-biomaterials science and supramolecular chemistry for producing desired stable assemblies from protein crystals,” concludes Ueno. Only time will tell what other useful molecular structures can be produced using this strategy and what interesting applications they will find!
###
Media Contact
Kazuhide Hasegawa
[email protected]
Related Journal Article
http://dx.