UNIVERSITY PARK, Pa. — The chemical steps in an important cellular modification process that adds a chemical tag to some RNAs have been revealed in a new study. Interfering with this process in humans can lead to neuronal diseases, diabetes, and cancers. A research team, led by chemists at Penn State, has imaged a protein that facilitates this RNA modification in bacteria, allowing the researchers to reconstruct the process. A paper describing the modification process appears Sept. 15 in the journal Nature.
Credit: Booker Lab, Penn State
UNIVERSITY PARK, Pa. — The chemical steps in an important cellular modification process that adds a chemical tag to some RNAs have been revealed in a new study. Interfering with this process in humans can lead to neuronal diseases, diabetes, and cancers. A research team, led by chemists at Penn State, has imaged a protein that facilitates this RNA modification in bacteria, allowing the researchers to reconstruct the process. A paper describing the modification process appears Sept. 15 in the journal Nature.
Transfer RNAs (tRNA) are the RNAs that “read” the genetic code and translate it into a sequence of amino acids to make a protein. The addition of a chemical tag — a methyl sulfur group — to a particular location on some tRNAs improves their ability to translate messenger RNA into proteins. When this modification process — called methylthiolation — doesn’t occur properly, mistakes can be incorporated into the resulting proteins, which in humans can lead to neuronal disease, cancer, and increased risk of developing Type 2 diabetes.
“Methylthiolation is ubiquitous across bacteria, plants, and animals,” said Squire Booker, a biochemist at Penn State and investigator with the Howard Hughes Medical Institute who led the research team. “In this study, we determined the structure of a protein called MiaB to better understand its role in facilitating this important modification process in bacteria.”
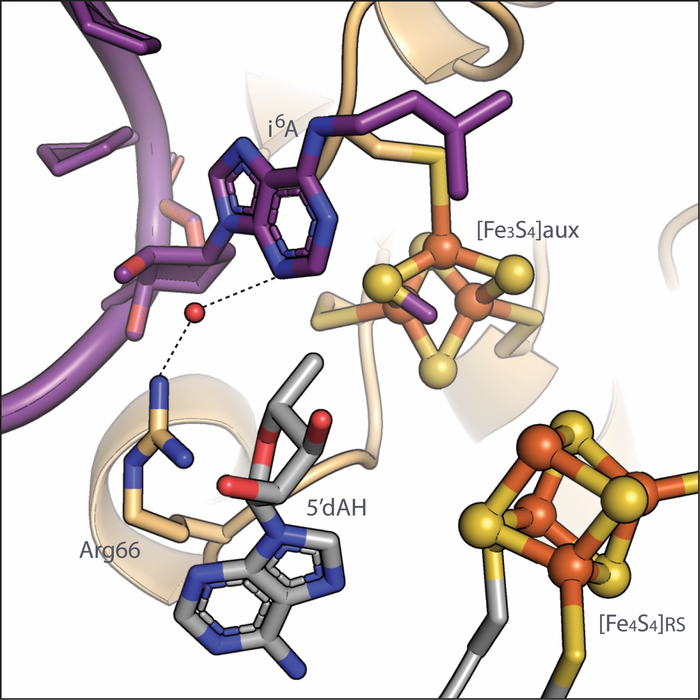
The MiaB protein from the bacteria Bacteroides uniforms is a member of the radical SAM (S-adenosylmethionine) family of enzymes. Radical SAM enzymes typically use one of their own iron-sulfur clusters to convert a SAM molecule into a “free radical” that helps move the reaction forward. Unlike most other radical SAM enzymes, MiaB contains two iron-sulfur clusters: a radical SAM cluster and an auxiliary cluster, where most of the intricate chemistry takes place.
Imaging MiaB in action with SAM molecules and tRNA at several points during methylthiolation allowed the researchers to infer the chemical steps during the modification process. First, a molecule of SAM donates its methyl group to the auxiliary iron-sulfur cluster on MiaB.
“The source of the sulfur atom attached to the tRNA has been controversial, but our structures reveal that a methyl group from SAM attaches to a sulfur atom on MiaB’s auxiliary iron-sulfur cluster,” said Olga Esakova, assistant research professor in chemistry at Penn State and first author of the paper. “This methyl group and the sulfur it attaches to on MiaB are ultimately what transfers to the tRNA, but some additional steps occur before the tRNA can accept the methylthio group.”
The addition of an electron fragments a second molecule of SAM into a free radical. The radical ultimately takes a hydrogen atom from the tRNA, which is replaced with the methylthio group on MiaB.
“Initially, the hydrogen on the tRNA is not positioned in a way that allows both access to the radical that removes it and access to the methylthio group that needs to be transferred, because the hydrogen and the atoms attached nearby are all aligned in the same plane,” said Booker. “Our structures show that the methylthio group on MiaB’s auxiliary cluster induces a change in geometry at that spot in the tRNA undergoing methylthiolation, which changes into more of a tetrahedral shape, with the hydrogen in an optimal position to be plucked off by the radical and the methylthio group in an optimal position for subsequent transfer.”
The result of these steps is tRNA with the added methylthio group and a successful modification.
Next, the researchers hope to identify how the auxiliary cluster is rebuilt after each turnover so that the process can proceed for multiple rounds. They are also investigating analogous proteins that play a similar role in the modification process in humans.
###
Booker is an Evan Pugh University Professor of Chemistry and of Biochemistry and Molecular Biology and Eberly Distinguished Family Chair in Science at Penn State. In addition to Booker and Esakova, the research team at Penn State includes Neela Yennawar, director of the Penn State X-Ray Crystallography and Automated Biological Calorimetry Core Facilities; Arthur Arcinas, a graduate student at the time of the research; Bo Wang, assistant research professor of chemistry; and Carsten Krebs, professor of chemistry and of biochemistry and molecular biology. The team also includes Tyler Grove and Steven Almo at the Albert Einstein College of Medicine.
This research was supported by the Howard Hughes Medical Institute, the National Institutes of Health, the National Science Foundation, the Penn State Eberly College of Science, The Price Family Foundation, and the Penn State Huck Institutes of the Life Sciences.
Journal
Nature
DOI
10.1038/s41586-021-03904-6
Method of Research
Imaging analysis
Subject of Research
Cells
Article Title
Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB
Article Publication Date
15-Sep-2021



