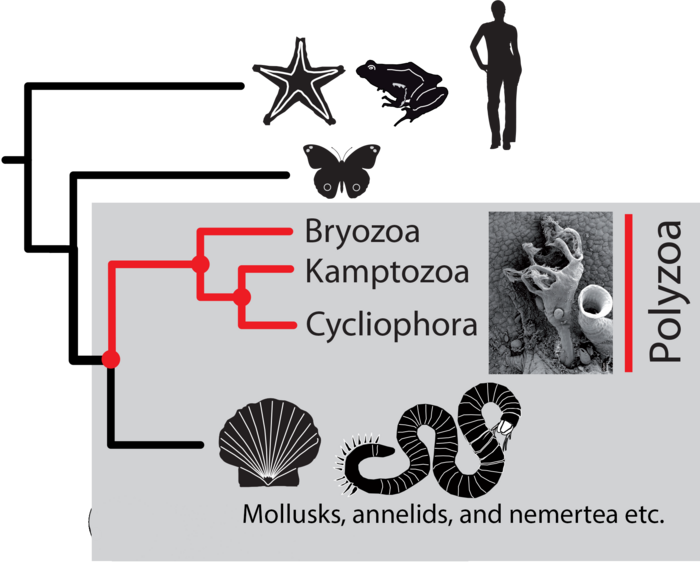
Kamptozoa and Bryozoa are two phyla of small aquatic invertebrates. They are related to snails and clams (collectively called mollusks), bristleworms, earthworms, and leeches (collectively called annelids), and ribbon worms (nemertea). But their precise position on the tree of life, and how closely related they are to these other animals, has always puzzled evolutionary biologists. Previous studies have consistently moved them around. What’s more, while Kamptozoa and Bryozoa were originally considered to form one group, they were separated based on their appearance and anatomy. Now, by using cutting-edge sequencing technology and powerful computational analysis, scientists from the Okinawa Institute of Science and Technology Graduate University (OIST), in collaboration with colleagues from St-Petersburg University and Tsukuba University, have revealed that the two phyla split from mollusks and worms earlier than previous studies have suggested, and thus they indeed form a distinct group.
Credit: OIST
- Kamptozoa and Bryozoa are two phyla of small aquatic invertebrates that are related to animals like snails, earthworms, leeches, and ribbon worms.
- But questions surrounding the precise location of these two phyla on the tree of life and how closely related they are to these other animals have always puzzled evolutionary biologists.
- To answer these questions, researchers sequenced the transcriptome of four species of Kamptozoa and two species of Bryozoa, but to a far higher quality level than had previously been achieved. While past datasets had completeness of 20-60%, in this study, the transcriptome completeness was over 96%.
- In doing so, the researchers revealed that the two phyla split from their relatives earlier than previous studies have suggested, and found that they form a distinct group, called Polyzoa.
- The dataset generated from this study could also be used to answer other fundamental evolutionary questions—such as the more precise evolutionary relationships of the other animals, and how life diversified.
Kamptozoa and Bryozoa are two phyla of small aquatic invertebrates. They are related to snails and clams (collectively called mollusks), bristleworms, earthworms, and leeches (collectively called annelids), and ribbon worms (nemertea). But their precise position on the tree of life, and how closely related they are to these other animals, has always puzzled evolutionary biologists. Previous studies have consistently moved them around. What’s more, while Kamptozoa and Bryozoa were originally considered to form one group, they were separated based on their appearance and anatomy. Now, by using cutting-edge sequencing technology and powerful computational analysis, scientists from the Okinawa Institute of Science and Technology Graduate University (OIST), in collaboration with colleagues from St-Petersburg University and Tsukuba University, have revealed that the two phyla split from mollusks and worms earlier than previous studies have suggested, and thus they indeed form a distinct group.
“We’ve shown that by using high quality transcriptomic data we can answer a long-standing question to the best of our current techniques,” said Dr. Konstantin Khalturin, Staff Scientist in OIST’s Marine Genomics Unit and first author of the paper published in Science Advances.
A genome is the full set of genetic information found in every cell. It is subdivided into genes. These genes are made up of DNA base pairs and each gene contains the instructions needed to create a protein, and thus leads to the proper care and maintenance of a cell. For the instructions to be carried out, the DNA must first be transcribed into RNA. A transcriptome is the result of this, like the reflection of a genome but written in RNA base pairs rather than DNA.
This genetic information differs among species. Those who are closely related have very similar genetic information, while a greater evolutionary distance results in more genetic differences. By using this data, researchers have improved our knowledge of animal evolution, but some questions still prove difficult to answer.
As Kamptozoa and Bryozoa are closely related to mollusks, annelids, and nemertea, small mistakes in the dataset, or missing data, can result in an incorrect placement on the evolutionary tree. Furthermore, while collecting these tiny animals, it’s easy to pick up other organisms, such as algae, that contaminate the sample. Dr. Khalturin highlighted that they were careful to avoid contamination and later screened their dataset for RNA of algae and small animals to remove any that might have come from them.
In total, the researchers sequenced the transcriptome of four species of Kamptozoa and two species of Bryozoa, but to a far higher quality level than had previously been achieved. While past datasets had completeness of 20-60%, in this study, the transcriptome completeness was over 96%.
Using these transcriptomes, they predicted proteins and compared them to similar data of 31 other species, some of which were closely related to Kamptozoa and Bryozoa, such as clams and bristleworms, and others which were more distant, such as frogs, starfish, insects, and jellyfish. The high-quality datasets meant that they could compare many different genes and proteins simultaneously. Dr. Khalturin credited the powerful computational capabilities that the researchers could access at OIST.
“Our main finding is that the two phyla belong together,” said Dr. Khalturin. “This result was originally proposed in the 19th century by biologists who were grouping animals based on what they looked like.”
While Dr. Khalturin stated that this question had now been answered to the best ability available, he also highlighted that the dataset could answer other fundamental evolutionary questions—such as the more precise location of mollusks and annelids on the tree of life, and how life diversified.
Journal
Science Advances
DOI
10.1126/sciadv.abo4400
Method of Research
Data/statistical analysis
Subject of Research
Animals
Article Title
Polyzoa is back: The effect of complete gene sets on the placement of Ectoprocta and Entoprocta
Article Publication Date
1-Jul-2022




