This study is led by Prof. Wei-Li Zhao (Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine).
Credit: ©Science China Press
This study is led by Prof. Wei-Li Zhao (Shanghai Institute of Hematology, State Key Laboratory of Medical Genomics, National Research Center for Translational Medicine at Shanghai, Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine).
G-protein coupled receptors (GPCRs) are critical targets for viral molecules and correlate with pathogenesis of multiple diseases including inflammation, auto-immune disorder, and cancer. Epstein-Barr virus (EBV) is the first-discovered human oncogenic virus and implicated in more than 10% of all pathogen-associated cancers. Despite the findings that persistent EBV infection shares strikingly similar immune evasion features as in EBV-driven cancers through recruiting immunosuppressive cells and activating immune checkpoints, how EBV interacts with GPCR signaling to induce disease pathogenesis needs further investigation.
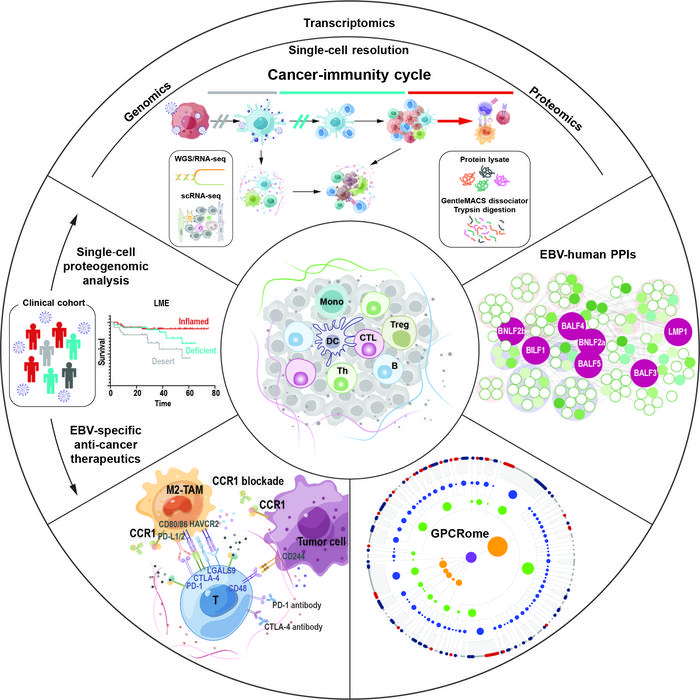
Natural killer T-cell lymphoma (NKTCL) is an aggressive lymphoid neoplasm in close association with EBV infection and thus represents a disease model to study EBV-induced lymphomagenesis. Jie Xiong et al. performed multi-omics analysis on 112 patients with NKTCL, integrating genomic (WGS), transcriptomic (RNA-seq and scRNA-seq) with mass spectrometry (MS)-based proteomics, depicted the lymphoma microenvironment (LME) at single-cell resolution and defined 3 different LME categories, namely immune-inflamed, -deficient, and -desert, corresponding to distinct setpoints of cancer-immunity cycle during EBV-host interactions and predicting clinical prognosis in the era of asparaginase-based targeted therapy.
To better understand the viral-cancer immunological interactions during oncogenesis, Jie Xiong et al. performed pull-down assay coupling mass spectrum to map the EBV-human protein-protein interaction network and identify proteins enriched in transcription regulation. ChIP-seq coupling RNA-seq further revealed that EBV protein transcriptional regulated GPCRs expression, which correlated with the LME categories. CCR1, highly expressed on EBV-positive lymphoma cells, regulated EBV DNA replications and gene expression, as well as malignant transformation. Besides, CCR1 was also detected in immunosuppressive M2-TAM and PMN/M-MDSC, correlating with the expression of immune checkpoints and contributing to the exhaustion of effective T cells through cell-cell contact. In consistent with the promising therapeutic effect of monoclonal antibody mogamulizumab, targeting another GPCR CCR4, in treating aggressive HTLV-1-driven adult T-cell leukemia-lymphoma, pre-clinical study demonstrated that CCR1 blockade not only directly killed tumor cells, but also enhanced cytotoxic cell expansion irrespective of molecular subtypes or LME categories.
In summary, as regulators of hematopoietic and immune cell trafficking, this study expanded the role of GPCRs as major regulators of the immune microenvironment in promoting tumor cell growth, determining immune phenotypes, and potentially serving as novel therapeutic targets in EBV-driven cancers.
Journal
Science Bulletin
DOI
10.1016/j.scib.2023.09.029