Epidemiological study revealed that Staphylococcus saprophyticus causing urinary tract infections can have origin in food
In young women, Staphylococcus saprophyticus is a main cause of urinary tract infections (UTI), reaching 20% prevalence. Understanding the epidemiology of this microorganism can help identify its origin, distribution, causes, and risk factors. Now, ITQB NOVA researchers led by Maria Miragaia showed evidence that Staphylococcus saprophyticus can originate in food, namely in the meat-production chain.
Europe is the world’s second-biggest producer of pork, the most favored meat type in these countries. One of the contaminants of that meat is S. saprophyticus, which is found also in the environment, the gut and rectal flora of pigs, and in the human gastrointestinal tract, vagina, and perineum.
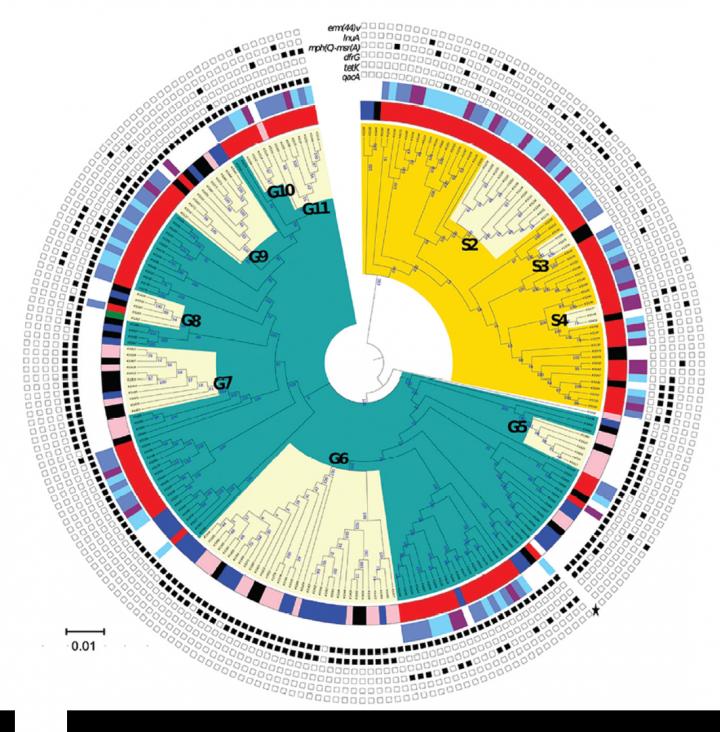
In the study published in the journal Emerging Infectious Diseases, by using a combination of phenotypic, genomic, and pan-genome wide association approaches, researchers identified two different lineages (G and S) of S. saprophyticus. The lineage G, from food origin and transmitted to humans by contact with food products, and lineage S, from human origin. Both are associated with disease and may be transmitted directly or indirectly between persons within the community, showing not only a local but an extensive geographic distribution.
In order to understand if these bacteria causing urinary tract infections could be related to the ones in pork, the research group looked at S. saprophyticus from a slaughterhouse and compared them to those causing human urinary tract infections. The team analysed bacteria collected from UTI worldwide over 20 years, and from UTI and pork meat production chain in Portugal.
The results revealed that bacteria from the slaughterhouse (equipment, meat, workers colonisation) were similar to human UTI bacteria and had the same antibiotic resistance profile. Although S. saprophyticus pig’s colonisation rate was extremely low (1%), 35% of slaughterhouse samples were contaminated. The presence of an antiseptic resistance gene (qacA) by all the lineage G bacteria could be part of the explanation for the ineffective cleaning procedures that were used. “S. saprophyticus strains of animal origin (lineage G) enters the slaughterhouse through food animals, persist on the equipment, disseminate and contaminate the meat processing chain and humans. Human colonization is a crucial step for the later occurrence of UTI”, explains Opeyemi Lawal, first author of the study developed during his PhD.
Additionally, by studying genomic data of bacteria collected from patients attending three hospitals in the Lisbon area, the researchers were able to clearly understand that the transmission of these pathogenic bacteria from both lineages (G and S) occurs between persons within the community. With this deep-structured analysis, researchers were also able to identify putative new virulence factors for this unexplored bacterium. The team will continue to search for reservoirs of this bacterium in humans and animals, and to study the mechanisms of S. saprophyticus dissemination and disease to provide the groundwork towards strategies to combat this pathogen, “This a clear example of how food manipulation can impact in human health, and how important it is to educate consumers regarding good individual hygiene practices to avoid spreading of infectious diseases”, says Maria Miragaia, head of the Bacterial Evolution and Molecular Epidemiology Lab. “This adds to the list of bacteria that are transmitted to humans through contact with animals and animal-derived food. But the exact mechanisms associated to the conversion from a colonizer to an infectious agent remains to be clarified”, adds Henrik Westh from the Copenhagen University Hospital – Amager and Hvidovre, University of Copenhagen (Denmark).
###
The research was developed in collaboration with ITQB NOVA PI Hermínia de Lencastre, Henrik Westh from Copenhagen University Hospital – Amager and Hvidovre, University of Copenhagen (Denmark) and Maria João Fraqueza, from the Centre for Interdisciplinary Research in Animal Health (Faculdade de Medicina Veterinária – Universidade de Lisboa), together with researchers and doctors from the Portuguese hospitals Hospital da Luz, Hospital Egas Moniz, and SAMS Hospital, and from the Hospital Universitari de Bellvitge (Spain) and the Narodowy Instytut Leków (Poland).
The work was supported by FCT (Fundação para a Ciência e Tecnologia), and performed within the projects funded by FEDER funds through COMPETE2020 – Programa Operacional Competitividade, funding MOSTMICRO Research Unit and by ONEIDA project co-funded by FEEI – “Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa2020”.
Media Contact
Renata Ramalho
[email protected]
Original Source
https:/
Related Journal Article
http://dx.