Credit: Palsson Lab
All life is subject to evolution in the form of mutations that change the DNA sequence of an organism’s offspring, after which natural selection allows the ‘fittest’ mutants to survive and pass on their genes to future generations. These mutations can generate new abilities in a species, but another common driving force for evolution is horizontal gene transfer (HGT) – the acquisition of DNA from a creature other than a parent, and even of a different species. For example, a significant amount of the human genome is actually viral DNA. Genetic engineering techniques now allow humans to intentionally induce HGT in various species to create ‘designer organisms’ capable of things like renewable chemical production, but it’s often difficult to get foreign DNA working in a new host.
Bioengineers at the University of California San Diego used genetic engineering and laboratory evolution to test the functionality of DNA placed into a new species and study how it can mutate to become functional if given sufficient evolutionary time. They published their results on August 10 in Nature Ecology and Evolution.
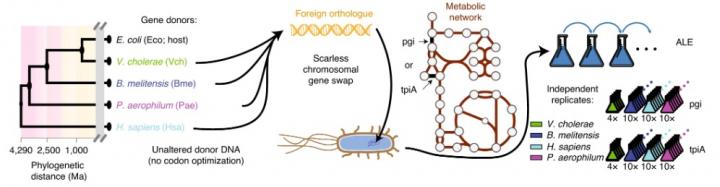
Using the model bacterium Escherichia coli as a host, bioengineers in Professor Bernhard Palsson’s Systems Biology Research Group used CRISPR to generate gene-swapped strains with donor DNA from species across the tree of life — from close bacterial relatives, to a microbe that lives in boiling hotsprings, to humans. The genes pgi or tpiA were replaced, two enzymes involved in sugar metabolism that cripple E. coli when removed, causing them to grow about 5 times slower. They then used an ‘evolution machine,’ robotic systems to study how the engineered bacteria adapted to replacement of such important genes with foreign versions. The automated systems enabled a large-scale study, generating hundreds of mutant strains evolved for more than 50,000 cumulative generations, something that would take decades rather than months if performed manually. Moreover, culture growth rates could be tracked in real-time as the populations evolved, allowing mutant strains to be isolated immediately after they took over the population from the ancestral strain. This high temporal resolution regularly allowed strains to be isolated that differed across their entire genome by only single mutations of interest, revealing not only order of acquisition but also providing an easy way to test the effect of mutations without laborious rounds of additional genetic engineering of the ancestral strains.
Although at first E. coli couldn’t use most of the foreign genes it was given, they quickly and frequently found an evolutionary way around this, often in a matter of days recovering from their crippled state to grow just as fast as before they were engineered. Notably, the foreign genes were not codon optimized before insertion into E. coli – this is regularly performed during synthetic HGT, relying on the fact that DNA codes for the string of amino acids that composes a protein via 3 letter codons that contain redundancy (e.g., Lysine is coded for by AAA or AAG). Different species have different genome-wide trends in codon usage that codon optimization minimizes for a gene inserted into a foreign species, but this was unnecessary to enable functionality – even for human DNA which has been evolutionarily diverging from E. coli for billions of years.
For every strain that successfully evolved use of the foreign DNA the critical factor was one or more mutations increasing gene expression level. Most of these mutations did not even occur within the foreign gene but rather in regions of E. coli‘s DNA controlling regulation of the gene, with their nature depending sensitively on the gene’s specific DNA sequence and location in the chromosome. Some of these mutations occurred with shocking regularity, including one observed independently more than 20 times, demonstrating that evolutionary outcomes can be (probabilistically) predicted to the single DNA basepair.
Of the few mutations occurring within the foreign DNA, most were at the beginning of the gene and ‘silent’ in nature, changing the codon but not the resulting amino acid. These are often assumed to have negligible impact on cell fitness (at least when in a single codon rather than across the entire gene as in codon optimization), but we found them to have significant impact. Thermodynamic modeling revealed that these mutations serve to prevent binding of the gene’s mRNA transcript into knotted structures, which limits the amount of protein that ribosomes can produce from the transcript. Finally, our hundreds of evolved strains contained >90 distinct mutations in the RNA Polymerase complex that produces mRNA transcripts from the DNA sequence. Such mutations are common in laboratory evolution experiments, but our large dataset revealed clustering of mutations into distinct regions depending on how the strain containing it evolved. This points to evolutionarily conserved regulatory strategies for rapidly adapting to metabolic perturbations such as the ones we induced.
“This result shows the importance of systems biology,” said UC San Diego bioengineering professor Bernhard Palsson, the principal investigator of the study. “Namely, biological function, in this case, is not so much about the parts of the cell, but how the parts come together to function as a system.”
Overall, this study establishes the influence of various DNA and protein features on cross-species genetic interchangeability and evolutionary outcomes, with implications for both natural HGT and strain design via genetic engineering.
###
This release was written by Troy Sandberg, first author of the paper.
Media Contact
Katherine Connor
[email protected]
Related Journal Article
http://dx.




