After replicating inside human cells, one foodborne pathogen uses a surprising pathway to get back out and infect new cells.
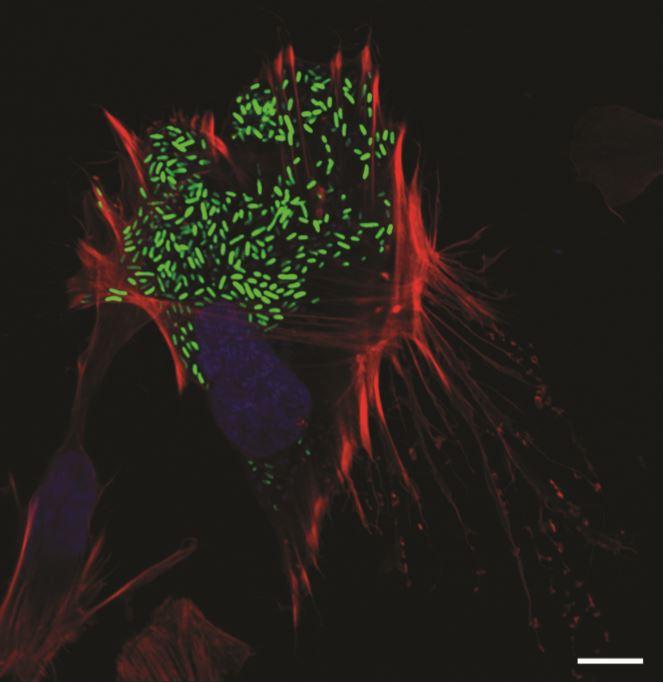
Credit: UT Southwestern Medical Center
DALLAS – Aug. 18, 2020 – As soon as the foodborne pathogen Vibrio parahaemolyticus infects a human intestinal cell, the bacteria are already planning their escape. After all, once it is in and multiplies, the bacterium must find a way out to infect new cells.
Now, UT Southwestern scientists have discovered the surprising route that V. parahaemolyticus takes during this exit – or egress – from cells. The bacteria, they report in the journal eLife, gradually modify cholesterol found in a cell’s plasma membrane, eventually weakening the membrane enough so that it can break through.
“The more we understand how bacteria are manipulating host cells at a molecular level, the more we understand how they cause disease,” says study leader Kim Orth, Ph.D., professor of molecular biology and biochemistry at UTSW and a Howard Hughes Medical Institute investigator. “Bacteria have many different mechanisms to escape, but this stood out because it’s an especially novel one.”
Vibrio bacteria are found in warm seawater and humans become infected by eating raw shellfish such as oysters. About a dozen different species of Vibrio can cause human illness; V. parahaemolyticus is the most common in the United States and leads to food poisoning symptoms – diarrhea, cramps, nausea, and vomiting.
About a decade ago, Orth’s group first revealed how V. parahaemolyticus infects human intestinal cells. Vibrio, they showed, uses a common bacterial system known as the type 3 secretion system 2 (T3SS2) to invade cells and begin replicating. The T3SS2 is composed of a large complex of proteins that form a needle that can inject molecules into a human cell, coaxing the cell to take in the bacteria and blocking any potential immune response.
“We started to get a good understanding of how this pathogen gets inside cells and maintains an existence,” says Orth. “We assumed that it was also using components of the T3SS2 to get out of cells again.”
But when Orth and her colleagues started studying the egress of V. parahaemolyticus out of human cells, the T3SS2 didn’t seem to play a role. Neither did a number of other known egress mechanisms that bacteria use. Finally, Marcela de Souza Santos – a former assistant professor of molecular biology at UTSW and co-first author of the study – suggested they search V. parahaemolyticus genome for proteins known as lipases, which can break down the fatty molecules that make up cellular membranes.
Orth’s team identified a lipase known as VPA0226 and thought they’d found their answer, assuming the lipase digested the membranes of human cells. But they were in for another surprise. When they tracked the activity of the lipase, they discovered that it instead headed for the mitochondria of cells, where it modified membrane cholesterol molecules. Over seven to eight hours, as these cholesterol molecules are modified, the cell membrane becomes weak. By this time, V. parahaemolyticus has multiplied – from one or two bacteria to about 500 – and all the copies can escape through the weakened membrane.
“This is the only report we know of where a bacterium uses this kind of T2SS lipase to egress from a host cell that was invaded in a T3SS2 dependent way,” says Suneeta Chimalapati, Ph.D., a research scientist in the Orth lab and co-first author of the study.
To confirm the role of VPA0226, de Souza Santos and Chimalapati tested what happened when V. parahaemolyticus completely lacked the lipase. Indeed, the bacteria successfully invaded human cells and began replicating, but remained stuck inside those initial cells. Eventually, the host cells – crammed full of bacteria – died along with all the V. parahaemolyticus.
The new observation likely won’t have any immediate therapeutic implications, the researchers say; V. parahaemolyticus usually resolves on its own without treatment. But it helps shed light on how bacteria evolve egress mechanisms and the importance of looking beyond known secretion systems when thinking about the important molecules used by bacterial pathogens.
“We really had tunnel vision thinking the T3SS2 dominated everything Vibrio did, but this shows how many other tools it has on hand to use for its pathogenesis,” says Orth, who holds the Earl A. Forsythe Chair in Biomedical Science and is a W.W. Caruth, Jr. Scholar in Biomedical Research. She was recently elected to the National Academy of Sciences.
###
Other UTSW researchers who contributed to this study were Alexander Lafrance, Ann Ray, Wan-Ru Lee, Giomar Rivera-Cancel, Goncalo Vale, Krzysztof Pawlowski, Matthew Mitsche, Jeffrey McDonald, and Jen Liou.
This research was supported by funds from the Howard Hughes Medical Institute, the National Institutes of Health (R01 GM115188, RO1 GM113079, PO1 HL20948), the Once Upon a Time Foundation, and The Welch Foundation (I-1561).
About UT Southwestern Medical Center
UT Southwestern, one of the premier academic medical centers in the nation, integrates pioneering biomedical research with exceptional clinical care and education. The institution’s faculty has received six Nobel Prizes, and includes 24 members of the National Academy of Sciences, 16 members of the National Academy of Medicine, and 13 Howard Hughes Medical Institute Investigators. The full-time faculty of more than 2,500 is responsible for groundbreaking medical advances and is committed to translating science-driven research quickly to new clinical treatments. UT Southwestern physicians provide care in about 80 specialties to more than 105,000 hospitalized patients, nearly 370,000 emergency room cases, and oversee approximately 3 million outpatient visits a year.
Media Contact
UT Southwestern Medical Center
[email protected]




