Credit: Kanazawa University
In the human body, proteins sometimes occur in fibrillar aggregates called amyloids. Although certain amyloids are known to have a biological function, amyloid formation is often associated with pathologies, including Alzheimer’s and Parkinson’s diseases. Understanding how exactly amyloid fibrils form is crucial for gaining insights into the development of such diseases and for advancing with treatment approaches. Now, Takahiro Watanabe-Nakayama from Kanazawa University, Kenjiro Ono from Showa University, and colleagues have investigated the formation process of particular amyloid fibrils using a technique enabling visualization of growth over time. The scientists specifically looked at the effect of cross-seeding (‘mixing’) different proteins forming aggregates, and found variations in elongation rates and the structures of the fibrils.
The researchers studied alpha-synuclein, a protein abundant in the human brain. They looked at what happened when letting wild-type alpha-synuclein molecules — the natural, most abundant variant — form aggregates, and also how aggregation is different when introducing (cross-seeding) mutant variants associated with Parkinson’s disease. In addition, the scientists examined the influence of the pH level of the microenvironment in which fibril growth takes place.
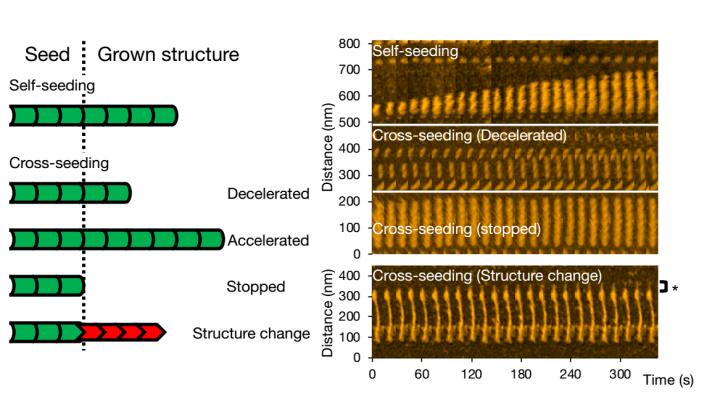
By means of high-speed atomic-force microscopy (HS-AFM), Watanabe-Nakayama, Ono and colleagues could record fibril aggregation at nanometer resolution and high video rate for various cases. First, the scientists looked at the growth of single variant types (self-seeding). They found that mutants produced more aggregates, or that they aggregated faster at neutral pH than the wild-type variants. Another observation was that elongation was faster at lower pH (5.8, i.e. acidic) than at higher pH (7.4, i.e. basic).
For cross-seeding, different scenarios can occur. Fibril growth can be accelerated or slowed down, or even stopped. The morphology of the original seed can be preserved, but it also happens that the structure of the resulting fibril is different — typical structural forms are ‘straight’ or ‘spiral’. The researchers checked that fibril structure and dynamics as observed with HS-AFM correspond to the processes in solution by means of fluorescence experiments; similar conclusions were obtained.
The findings of Watanabe-Nakayama, Ono and colleagues are relevant for better understanding amyloid-related diseases. Quoting the researchers: “Cross-seeding combined with variations in elongation rates has the effect of increasing the structural diversity of the resulting assemblies. This diversity may be reflected in distinct neurotoxic effects for various [protein] assemblies.”
###
[Background]
Amyloids
Amyloids are fibrillar aggregates of protein molecules. Although some amyloids are known to have a biological function (for example the release of hormones), they are associated with diseases collectively called amyloidoses. These include neurodegenerative disorders like Alzheimer’s and Parkinson’s diseases. Many different proteins can form amyloids; Takahiro Watanabe-Nakayama from Kanazawa University and colleagues studied fibril formation of alpha-synuclein molecules by means of high-speed atomic force microscopy.
Atomic force microscopy
Atomic force microscopy (AFM) is an imaging technique in which the image is formed by scanning a surface with a very small tip. Horizontal scanning motion of the tip is controlled via piezoelectric elements, while vertical motion is converted into a height profile, resulting in a height distribution of the sample’s surface. As the technique does not involve lenses, its resolution is not restricted by the so-called diffraction limit. In a high-speed setup (HS-AFM), the method can be used to produce movies of a sample’s structural evolution in real time. Watanabe-Nakayama and colleagues have successfully used HS-AFM to study the formation and structural dynamics of amyloids obtained by self- and cross-seeding alpha-synuclein protein variants.
Media Contact
Hiroe Yoneda
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




