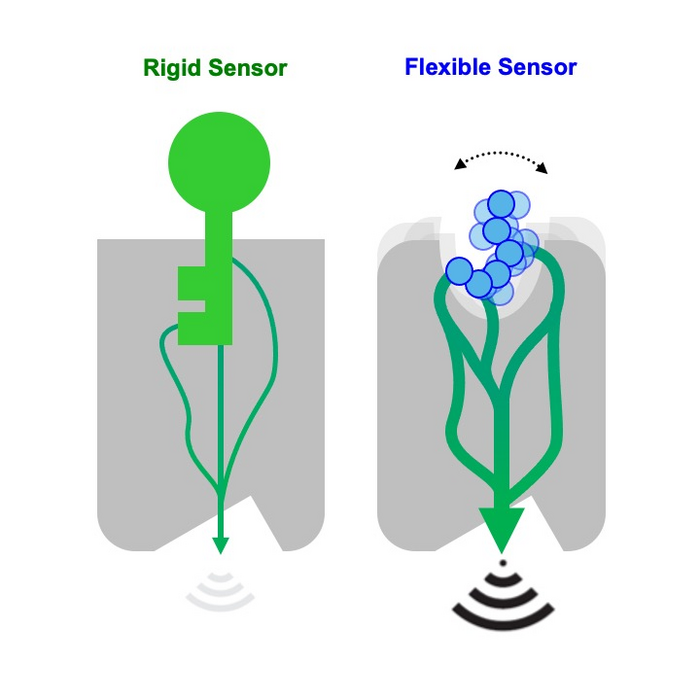
Biosensors are artificial molecular complexes designed to detect the presence of target chemicals or even biomolecules. Consequently, biosensors have become important in diagnostics and synthetic cell biology. However, typical methods for engineering biosensors focus on optimizing the interactions between static binding surfaces, and current biosensor designs can only recognize structurally well-defined molecules, which can be too rigid for “real-life” biology.
Credit: Robert E. Jefferson (EPFL)
Biosensors are artificial molecular complexes designed to detect the presence of target chemicals or even biomolecules. Consequently, biosensors have become important in diagnostics and synthetic cell biology. However, typical methods for engineering biosensors focus on optimizing the interactions between static binding surfaces, and current biosensor designs can only recognize structurally well-defined molecules, which can be too rigid for “real-life” biology.
“We developed a novel computational approach for designing protein-peptide ligand binding and applied it to engineer cell-surface chemotactic receptors that reprogrammed cell migration,” says EPFL professor Patrick Barth. “We think that our work could broadly impact the design of protein binding and cell engineering applications.”
The new biosensors developed by Barth’s group can sense flexible compounds and trigger complex cellular responses, which open up new possibilities for biosensor applications. The researchers created a ‘computational framework’, which is a computer-based system, for designing protein complexes that can change their shape and function dynamically – as opposed to the conventional static approaches. The framework can look at previously unexplored protein sequences to come up with new ways for the protein’s groups to be activated, even in ways that are different to their natural function.
The researchers used their new method to create synthetic receptors that can sense and respond to multiple natural or engineered molecular signals, providing optimal sensing of flexible ligands and strong allosteric signaling responses, a term that refers to changes in protein activity when a molecule binds to a different site on a protein, causing a change in the protein’s shape and activity at a different site.
The designed receptors act by interacting with the flexible ligands via allosteric triggers, like natural receptors, but they improve and rearrange how the signals are transferred, a bit like dialing the same number from a different cell phone with better service. Specifically, the triggers seem to funnel signals through the same set of “transmission hubs” as the natural ones, but considerably enhance signal transmission through optimally rewired dynamic couplings.
The research shows that combining a flexible sensing layer with a robust signal transmission layer may be a common hallmark of G protein-coupled receptors, a family of enormously important receptors in the cell, connected to virtually every major aspect of its life and function.
“We were able to leverage our biosensor design to drive cell migration in lymphocytes, which migrate more efficiently towards chemokines when equipped with designed biosensors,” says Rob Jefferson, the study’s first author. “Chemokines serve as chemical beacons for immune cell recruitment in the body, a suboptimal process in certain diseases that could be improved with our biosensors.”
The new method of designing synthetic receptors could be useful in a wide variety of therapeutic contexts. For example, engineered cytotoxic lymphocytes with enhanced chemotaxis toward tumor sites could prove useful in cancer treatment. Designing receptors that can sense and respond to specific signals, provides a promising new synthetic cell biology tool, leading to more precise control over cellular processes for a wide range of therapeutic applications.
Reference
Jefferson RE, Oggier A, Fuglistaler A, Camviel N, Hijazi M, Rico Villarreal A, Arber C, Barth P. Computational design of dynamic receptor:peptide signaling complexes applied to chemotaxis. Nature Communications 14, 2875. 19 May 2023. DOI: 10.1038/s41467-023-38491-9
Journal
Nature Communications
DOI
10.1038/s41467-023-38491-9



