A deep learning-based tool predicts transcription factors using protein sequences as inputs
Credit: KAIST
A joint research team from KAIST and UCSD has developed a deep neural network named DeepTFactor that predicts transcription factors from protein sequences. DeepTFactor will serve as a useful tool for understanding the regulatory systems of organisms, accelerating the use of deep learning for solving biological problems.
A transcription factor is a protein that specifically binds to DNA sequences to control the transcription initiation. Analyzing transcriptional regulation enables the understanding of how organisms control gene expression in response to genetic or environmental changes. In this regard, finding the transcription factor of an organism is the first step in the analysis of the transcriptional regulatory system of an organism.
Previously, transcription factors have been predicted by analyzing sequence homology with already characterized transcription factors or by data-driven approaches such as machine learning. Conventional machine learning models require a rigorous feature selection process that relies on domain expertise such as calculating the physicochemical properties of molecules or analyzing the homology of biological sequences. Meanwhile, deep learning can inherently learn latent features for the specific task.
A joint research team comprised of Ph.D. candidate Gi Bae Kim and Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering at KAIST, and Ye Gao and Professor Bernhard O. Palsson of the Department of Biochemical Engineering at UCSD reported a deep learning-based tool for the prediction of transcription factors. Their research paper “DeepTFactor: A deep learning-based tool for the prediction of transcription factors” was published online in PNAS.
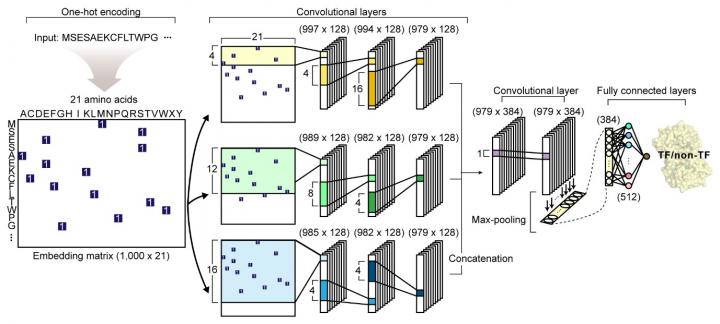
Their article reports the development of DeepTFactor, a deep learning-based tool that predicts whether a given protein sequence is a transcription factor using three parallel convolutional neural networks. The joint research team predicted 332 transcription factors of Escherichia coli K-12 MG1655 using DeepTFactor and the performance of DeepTFactor by experimentally confirming the genome-wide binding sites of three predicted transcription factors (YqhC, YiaU, and YahB).
The joint research team further used a saliency method to understand the reasoning process of DeepTFactor. The researchers confirmed that even though information on the DNA binding domains of the transcription factor was not explicitly given the training process, DeepTFactor implicitly learned and used them for prediction. Unlike previous transcription factor prediction tools that were developed only for protein sequences of specific organisms, DeepTFactor is expected to be used in the analysis of the transcription systems of all organisms at a high level of performance.
Distinguished Professor Sang Yup Lee said, “DeepTFactor can be used to discover unknown transcription factors from numerous protein sequences that have not yet been characterized. It is expected that DeepTFactor will serve as an important tool for analyzing the regulatory systems of organisms of interest.”
###
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation (NRF) of Korea.
About KAIST
KAIST is the first and top science and technology university in Korea. KAIST was established in 1971 by the Korean government to educate scientists and engineers committed to the industrialization and economic growth of Korea.
Since then, KAIST and its 64,739 graduates have been the gateway to advanced science and technology, innovation, and entrepreneurship. KAIST has emerged as one of the most innovative universities with more than 10,000 students enrolled in five colleges and seven schools including 1,039 international students from 90 countries.
On the precipice of its semi-centennial anniversary in 2021, KAIST continues to strive to make the world better through the pursuit in education, research, entrepreneurship, and globalization.
Media Contact
Younghye Cho
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




