In the world of biological research, understanding the spatial arrangement of cells within tissues is crucial for deciphering the complexities of cellular interactions and disease pathogenesis. Recent advancements in spatial transcriptomics techniques have allowed scientists to map gene expression across tissues while preserving their structural integrity. These developments are crucial in the context of exploring healthy and diseased states of biological tissues, especially in light of diseases like cancer.
However, despite the advancements, researchers face significant challenges in accurately identifying spatial domains within tissues based on gene activity. Traditional approaches often fall short as they employ arbitrary distance parameters that may not align with the biological boundaries present in complex tissues. Some methods attempt to enhance accuracy by incorporating multiple tissue images; yet, they are often hampered by inconsistencies in image quality or the availability of data, necessitating cumbersome manual adjustments and alignment processes that can lead to errors and inefficiencies.
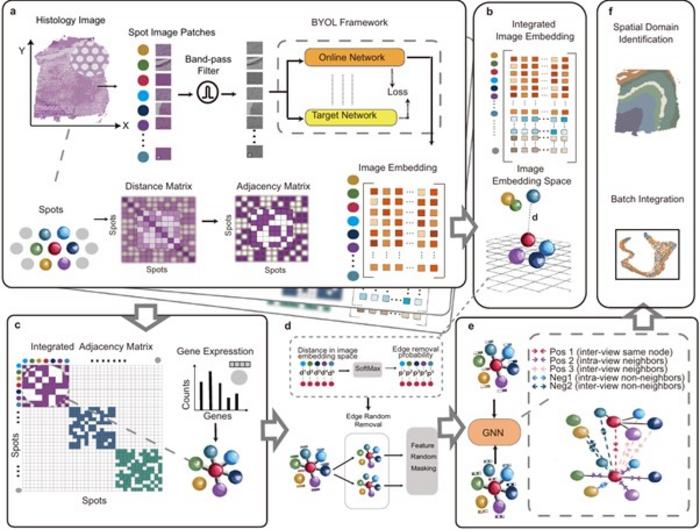
The STAIG framework exemplifies a new approach to processing histological images through segmentation into small patches. By employing self-supervised learning models, STAIG extracts relevant features from these patches in a way that eliminates the need for extensive pre-training, thus streamlining the analysis process. Ultimately, STAIG constructs a strategic graph structure where nodes represent gene expression data while edges indicate spatial relationships, effectively managing vertically stacked images.
During rigorous benchmark evaluations, STAIG was compared to other leading-edge spatial transcriptomics techniques, revealing superior performance across varied conditions, particularly in scenarios where spatial alignment was unavailable or histological images were absent. In datasets relating to human breast cancer and zebrafish melanoma, STAIG showcased exceptional acuity in recognizing spatial regions, including those complex areas that previously resisted detection by existing methodologies. The precision in delineating tumor boundaries and transitional zones underlines STAIG’s applicability and potential to advance cancer research significantly.
What sets STAIG apart is its foundation in deep-learning principles, which are increasingly gaining traction in the biological field. By leveraging robust model architecture and supplemental image data, STAIG ensures high accuracy in spatial domain identification. The implications of this framework extend beyond cancer studies, opening doors to potential applications across a diverse range of biological investigations.
Professor Nakai and his team hold tremendous optimism regarding the STAIG framework and its future applications, particularly in the realms of medical research and biology. As Nakai points out, the implementation of STAIG can greatly expedite the analysis of spatial transcriptome data, bringing new clarity to the intricate structures of biological systems. This includes vital explorations into the interactions between cancer cells and their surrounding environments, as well as insights into organ formation during embryonic development.
The promise of STAIG lies not only in its methodological superiority but also in the potential it holds for transforming our understanding of complex biological mechanisms. As research in this field continues to evolve, scientists expect that the insights gleaned through spatial transcriptomics will deepen our comprehension of fundamental processes underlying health and disease, ultimately guiding the development of novel therapeutic interventions for a myriad of illnesses.
Further studies and explorations centered around STAIG will ensure that the vast potential of spatial transcriptomics is fully realized, paving the way for breakthroughs that can redefine our approach to biological research and subsequently enhance our overall understanding of health and disease.
As the scientific community embraces the revolutionary capabilities of STAIG, it anticipates that this integrated approach will not only improve the accuracy of spatial domain identification in tissues but also remedy the longstanding challenges preceding translational medicine. The future of spatial transcriptomics is undeniably bright, and with continued investment and research, the full spectrum of its applications will soon come to light, elevating the frontiers of science.
Subject of Research: Human tissue samples
Article Title: STAIG: Spatial transcriptomics analysis via image-aided graph contrastive learning for domain exploration and alignment-free integration
News Publication Date: 27-Jan-2025
Web References: Nature Communications Paper
References: Kenta Nakai et al.
Image Credits: Professor Kenta Nakai, Institute of Medical Science, The University of Tokyo, Japan
Keywords: Spatial transcriptomics, deep learning, cancer research, gene expression, histological images, graph contrastive learning, biological systems.
Tags: cancer research advancementscellular interactions in tissueschallenges in spatial domain identificationdeep learning frameworks in biologygene expression mappingimage quality issues in researchinnovative frameworks in biomedical researchmanual adjustments in data analysisNature Communications publicationsProfessor Kenta Nakai contributionsspatial transcriptomics advancementstissue analysis techniques