Credit: Earlham Institute
Throughout evolutionary history, there have been genetic elements that have duplicated – giving rise to genes with different functions. These are called 'paralogs'. They are able to form and evolve new functions, which have similar functions in relation to cellular signalling. This also means that there are many duplicated genes within the genome that might be redundant or less prominent when it comes to key cellular signalling pathways.
Scientists in the UK and Hungary, led by Earlham Institute (EI), have discovered which proteins are critical in a range of biological functions, including cellular communication.
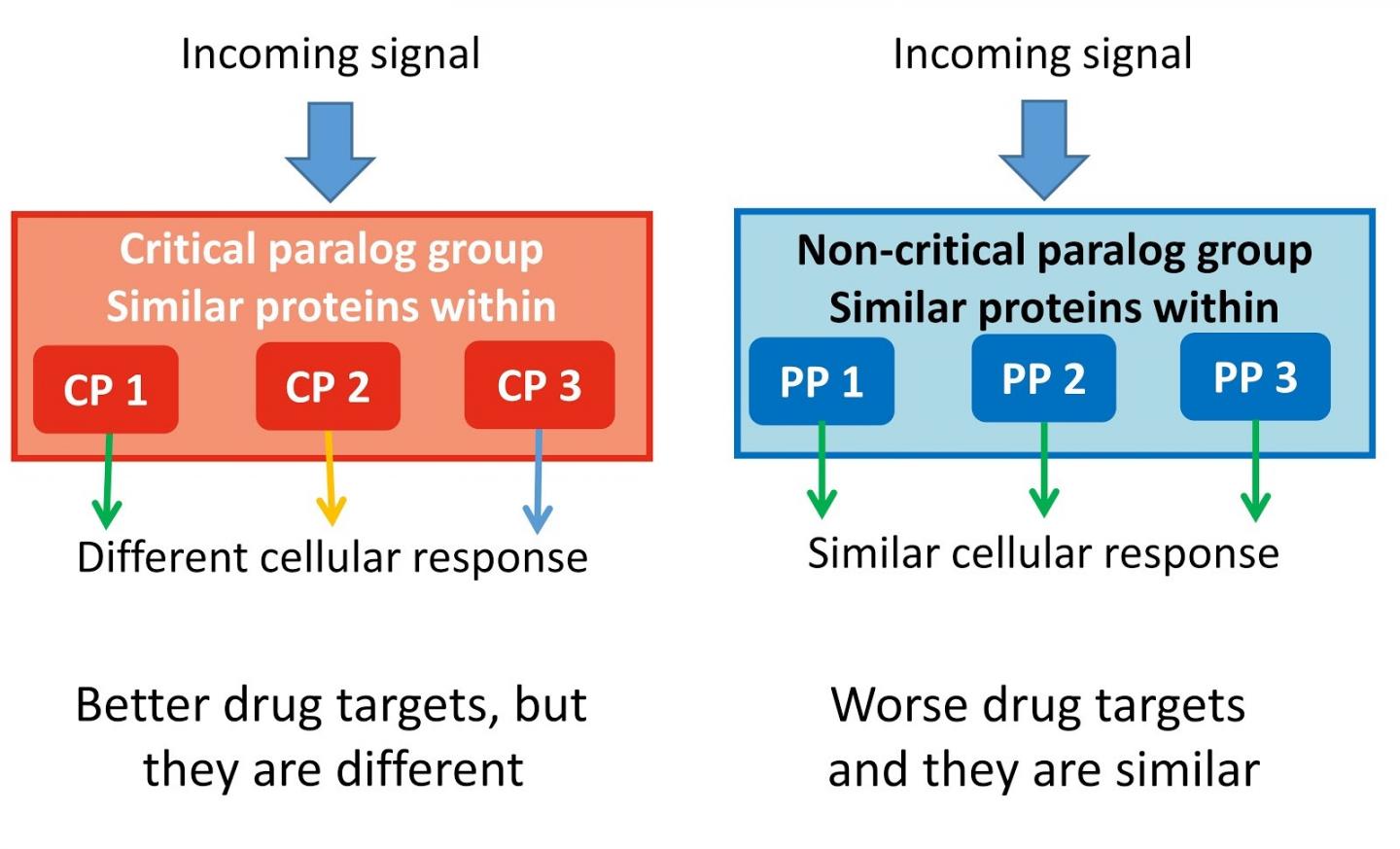
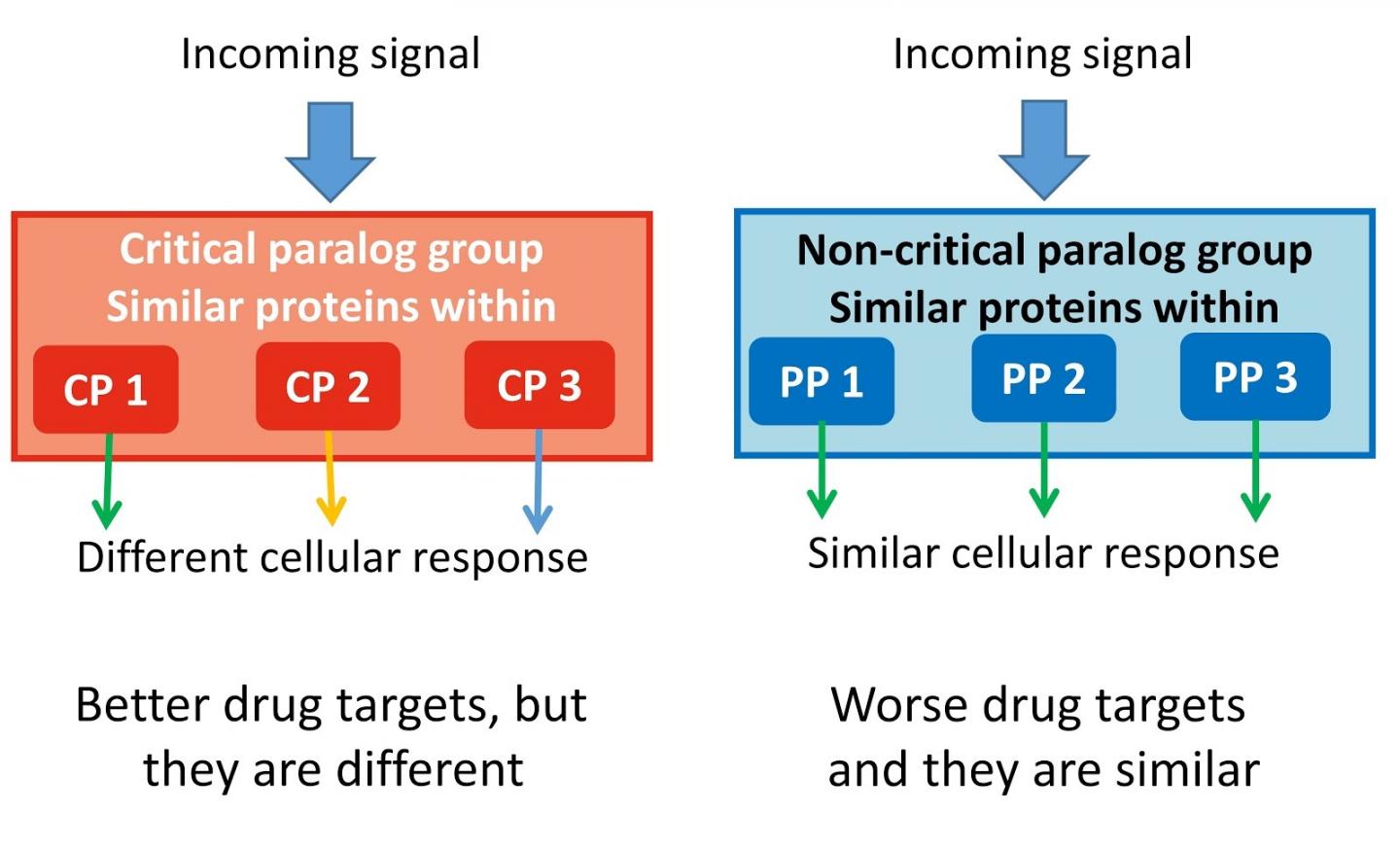
The team found 75 so-called critical paralog groups (CPGs) containing proteins with close evolutionary relationship to each other. One or two members in these groups can be critically important for a specific function, and also their changes, called mutations, can cause cancer or other inherited diseases.
The systematic discovery of these proteins identifies their indispensable role in human cellular signalling pathways, as well as how to potentially guide drug targets and find biomarkers for disease diagnosis in the future.
Lead author Tamas Korcsmaros, Fellow at EI and the Institute of Food Research, said: "Our cells must be able to detect and respond to many different pieces of genetic information coming from both internal and external sources. However, not all proteins in the cell are equally important.
In the post-genomic era, we already know there are key groups of proteins responsible for detecting, transmitting and communicating through cross-talk between different cellular pathways. Previously, it was challenging to identify which proteins, in which group, are more critical for the overall function of the cell; and those that are more relevant in causing diseases such as cancer, diabetes or neurodegenerative disorders.
The computational biology workflow we developed and confirmed with known examples provides an easily applicable method for disease-specific analysis. The concept will also help us to understand fundamental biological questions in comparative genomics on how duplication in the course of evolution led to more complex organs (such as the brain) and organisms."
First author Dezs? Módos, Research Associate at the University of Cambridge, added: "Our study shows the importance of similar proteins (paralogs) in signalling networks. Taking into account the paralog specificity in drug discovery because different paralog-specific signal routes could lead to totally different results like cell death or proliferation."
###
The paper titled: "Identification of critical paralog groups with indispensable roles in the regulation of signalling flow" is published in Scientific Reports. DOI:10.1038/srep38588.
Media Contact
Hayley London
[email protected]
07-760-438-218
http://www.earlham.ac.uk/
############
Story Source: Materials provided by Scienmag