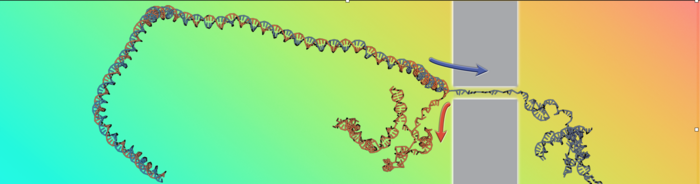
Reconstructing accurately how the parts of a complex molecular are held together knowing only how the molecule distorts and breaks up. This was the challenge taken on by a research team led by SISSA’s Cristian Micheletti and recently published on Physical Review Letters. In particular, the scientists studied how a DNA double helix unzips when translocated at high velocity through a nanopore, reconstructing fundamental DNA thermodynamic properties from the sole speed of the process.
Credit: Antonio Suma and Cristian Micheletti
Reconstructing accurately how the parts of a complex molecular are held together knowing only how the molecule distorts and breaks up. This was the challenge taken on by a research team led by SISSA’s Cristian Micheletti and recently published on Physical Review Letters. In particular, the scientists studied how a DNA double helix unzips when translocated at high velocity through a nanopore, reconstructing fundamental DNA thermodynamic properties from the sole speed of the process.
The translocation of polymers through nanopores has long studied as a fundamental theoretical problem as well as for its several practical ramifications, e.g. for genome sequencing. We recall that the latter involves driving a DNA filament through a pore so narrow that only one of the double-helical strands can pass, while the other strand is left behind. As a result, the translocated DNA double helix will necessarily split and unwind, an effect known as unzipping.
The research team, which also includes Antonio Suma from the University of Bari, first author, and Vincenzo Carnevale from Temple University, used a cluster of computers to simulate the process with different driving forces keeping track of the DNA’s unzipping speed, a type of data that has rarely been studied despite being directly accessible in experiments. Using previously developed theoretical and mathematical models, researchers were able to work “backwards”, using the information on the speed to accurately reconstruct the thermodynamics of the formation and rupture of the double-helix structure.
“Previous theories”, the researchers explain, “set off from detailed knowledge of the thermodynamics of a molecular system which was then used to predict the response to more or less invasive external stresses. This alone is a major challenge in itself. We looked at the inverse problem: we started from the DNA’s response to aggressive stresses, such as the forced unzipping of the double helix, to recover the details of the thermodynamics. Due to the invasive and rapid nature of the unzipping process, the project seemed doomed to fail, and that was probably why it had never been tried before. However, we also knew that the right theoretical and mathematical models, if applicable, could offer us a promising solution to the problem. After analysing the extensive set of collected data, we were very thrilled to discover that this was exactly the case; we were happy we had the right intuition.”
The technique adopted in the study is general, and thus the researchers expect to be able to extend it beyond DNA to other molecular systems that are still relatively unexplored. A case in point are the so-called molecular motors, protein aggregates that use energy to make cyclic transformations, very much like the engines in our everyday life. “Up until now”, researchers stress, “studies on molecular motors have started by formulating hypothesises on their thermodynamics and then comparing predictions with experimental data. The new method that we have validated should allow taking the inverse route, namely using data from out-of-equilibrium experiments to recover the thermodynamics, with clear conceptual and practical advantages.”
Journal
Physical Review Letters
DOI
10.1103/PhysRevLett.130.048101
Method of Research
Computational simulation/modeling
Subject of Research
Not applicable
Article Title
Nonequilibrium Thermodynamics of DNA Nanopore Unzipping
Article Publication Date
27-Jan-2023




