Study conducted in living cells opens up new opportunities for the development of more effective therapies against different types of cancer, such as glioblastoma
Credit: Guilherme de Oliveira
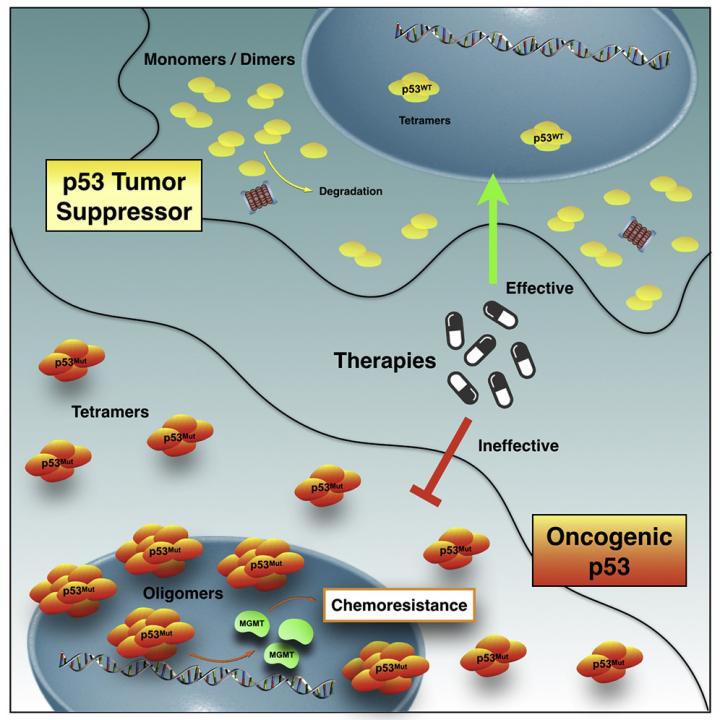
More than half of cancer cases worldwide are associated with genetic mutations in p53, the protein responsible for protecting DNA from changes that can lead to cancer. When this protein deforms, it not only loses its protective capacity, but can also gain new functions, acting as a “traitor”, and contributing to the spread of the tumor by forming protein clusters that may be resistant to chemotherapy. The mechanisms by which this “betrayal” occurs and how it causes drug resistance are not yet fully understood.
A group of Brazilian researchers led by biochemist Jerson Lima Silva, professor at the Federal University of Rio de Janeiro (UFRJ), uncovered part of the mystery. In a new study, the scientists identified the presence of a large amount of the protein’s traitor version in chemotherapy-resistant cells derived from glioblastoma, a super aggressive brain tumor. They also found how the deformed protein are organized inside the cell to exert resistance: they form clumps larger than those found in healthy individuals, some with amyloid properties, that is, when mutation leads to clumps. These structures were observed in the nucleus of living cells for the first time. The results are published in iScience.
The use of living cells makes important contributions to research in the field. “Unlike other studies, we identified small oligomers (structures a little larger than the healthy version of p53) in living cells, contributing to the identification of p53 aggregates more closely to what should occur in the human organism. This was possible because of the collaboration with researcher Enrico Gratton, who developed fluorescence fluctuation spectroscopy analysis,” explains Murilo Pedrote, a graduate student at the National Institute of Science and Technology for Structural Biology and Bioimaging (INBEB) and first author of the study.
The research was conducted using a p53-specific mutation (M237I). “This is important because the native protein (without mutation) and other mutations are not capable of conferring the same drug resistance,” says Guilherme A. P. de Oliveira, professor at UFRJ and one of the study’s coordinators.
Lima Silva’s group has been studying p53 mutations for over 15 years. His laboratory was the first to identify the propensity of the deformed protein to form amyloid aggregates, and that they play a crucial role in cancer development through loss of function, negative dominance (when mutated versions of the protein bind to healthy ones, altering their behavior) and gain of function. After that, it was observed by different scientists that mutated p53 amyloid aggregates are present in breast, ovarian and prostate cancer. Due to their characteristics, these amyloid clusters have become a new target for anticancer therapy.
In the new study, Lima Silva, de Oliveira and the other authors found that, in glioblastoma, an aggressive, invasive brain tumor with low survival (14 months), the deformed protein is involved not only in the formation of amyloid aggregates, but also in the resistance to temozolomide, the main drug used to treat the disease. The results are expected to enable the development of more effective treatments against various types of cancer. “Our study indicates that misfolded p53 and p53 aggregates are formidable targets for the development of new cancer treatment strategies,” Lima Silva concludes.
###
The paper entitled “Oncogenic gain of function in glioblastoma is linked to mutant p53 amyloid oligomers” is published online in iScience.
The study was funded by the National Council for Scientific and Technological Development (CNPq), the Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ), the Coordination of Improvement of Higher Education (CAPES / MEC) and the National Institute of Science and Technology for Structural Biology and Bioimaging (INBEB).
Media Contact
Jerson Lima Silva
[email protected]
55-219-993-90502
Original Source
https:/
Related Journal Article
http://dx.




