Credit: Image courtesy of Rice University
Elaborate molecular networks inside living cells enable them to sense and process many signals from the environment to perform desired cellular functions. Synthetic biologists have been able to reconstruct and mimic simpler forms of this cellular signal processing. But now, a new toolset powered by self-assembling molecules and predictive modeling will allow researchers to construct the complex computation and signal processing found in eukaryotic organisms, including human cells.
The work by Assistant Professor Ahmad ‘Mo’ Khalil (BME), Assistant Professor Caleb Bashor of Rice University, BME graduate student Nikit Patel, and other colleagues at MIT, Harvard, the Broad Institute and Brandeis University, has been published in Science.
The type of combinatorial signal processing that they’ve been able to engineer synthetically is what cells naturally and elegantly do to enable intricate tasks, like those in embryonic development and differentiation.
“This work is a tour-de-force of synthetic biology that addresses a major question in how cells process information at the DNA level,” says Dr. Tom Ellis, a Reader in the Department of Bioengineering at Imperial College London, who was not involved in the study.
“By taking a common principle that we know exists in nature, the ability for regulatory molecules to collaborate and form higher-order assemblies, you can program cells to execute very difficult computational and combinatorial problems,” says Khalil. “This represents a very different way of engineering genetic circuits than is traditionally done and it can open up a new class of cellular functions that we can mimic and control.”
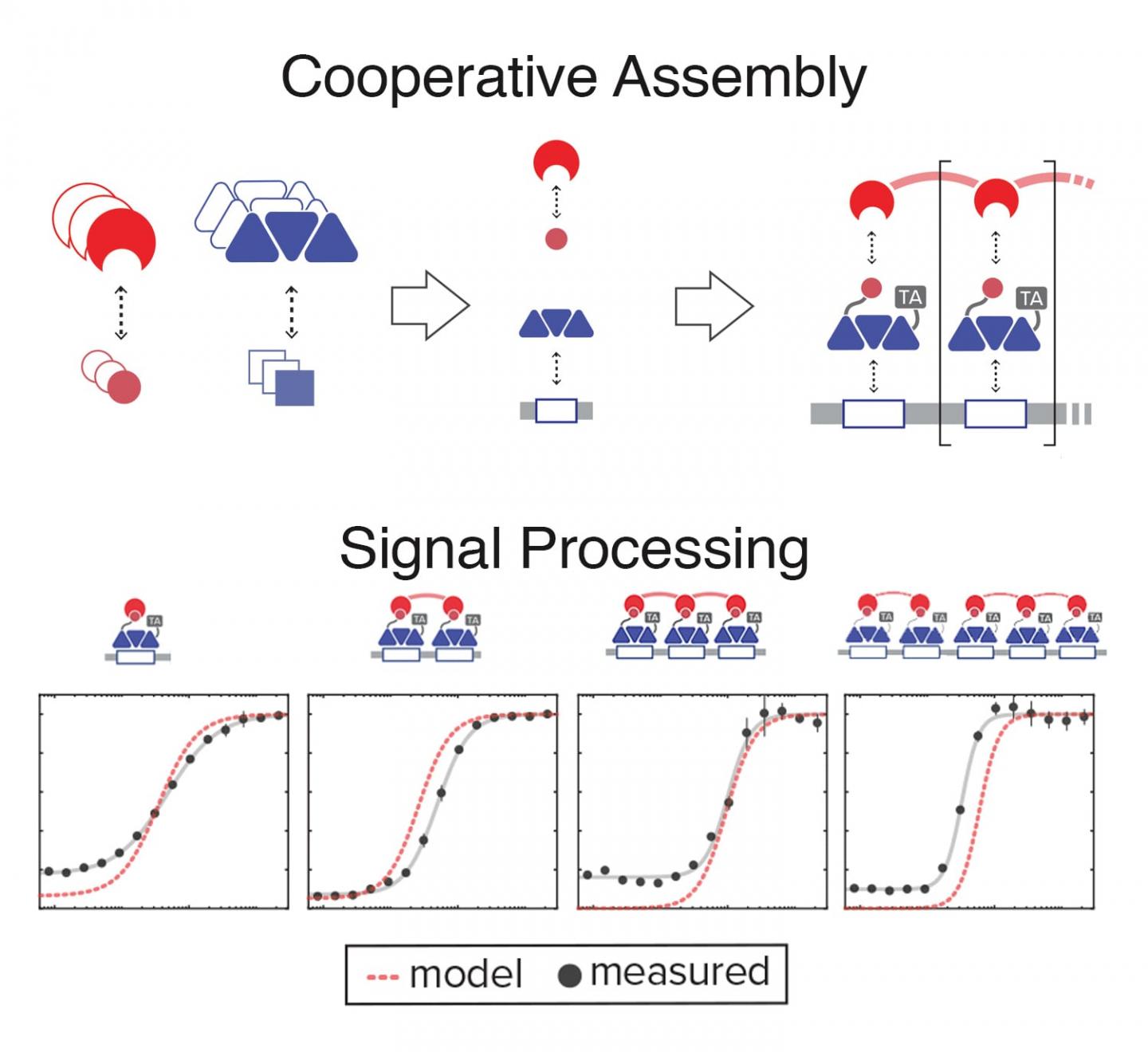
First, the researchers built a library of simple, synthetic molecular components that can interact with one another. Each of these components has its own unique chemical and kinetic makeup, which can be used to understand its behavior. Using these known properties, they constructed a quantitative model that can predict how different combinations of those molecules come together to build higher-order assemblies. They could then use that predictive model as a guide to design genetic circuits that take advantage of combinatorial assembly to execute desired signal processing functions.
“Basically, these components bind to one another with extremely weak interactions,” says Bashor. “But all of those weak interactions add up, in a bigger complex, to something that’s really tight. So, when there’s very few of them floating around, they won’t form the complex. And when they reach a critical concentration, they see each other, and they can basically come together and form the complex.”
The complex itself it made up of three components: a synthetic transcription factor that controls gene activation, the sequence of DNA where that transcription factor binds, and a synthetic “clamp” molecule that secures the three pieces together. That complex can allow them to tune the strength of the cell’s response to an input signal, and shut on and off the response at desired times. But it’s also much more than that.
“What we are harnessing and trying to build is one of the most powerful and universal features of biology: cooperativity,” says Khalil. “One way to think about cooperativity is that it allows for the whole to be greater than the sum of its parts.”
“You can think about cooperativity as the same type of signal-processing feature that gives you an analog-to-digital converter,” says Bashor. “An analog-to-digital converter takes something that’s basically linear and turns it into something switch-like.”
Using their system, engineered cells are made to produce assembly components in response to a desired chemical or environmental input. In one experiment, they programmed yeast cells to respond to two different chemicals, and specifically to respond to varying concentrations of the two chemicals in either analog or digital fashion.
In the analog circuits, the response is continuous; if a small concentration of either or both chemicals is present, there is a graded response. But in the digital circuits, there is an all-or-nothing, discrete response–like a signal conversion to binary code, which is only comprised of 0s and 1s, off and on.
By taking advantage of their new ability to adjust the cooperativity between assembly components, they showed that they could convert the cell’s response from dull to sharp–the more cooperative the complex, the sharper the response. The sharpness of a response–how acutely and intensely a system responds when a signal reaches a critical threshold–is key for digital signal processing.
“Engineering this type of response into transcription factors was central for allowing us to program cells to perform a diverse array of complex functions, such as Boolean logic, time-dependent filtering, and even frequency decoding,” says Khalil.
That upgrade from analog to digital is the culmination of years of research. The analog-to-digital converter and their other synthetic gene circuits can be used to explore and manipulate the regulatory programs that guide immune and stem cell functions with the ultimate goal of developing transformational cell-based therapeutics from engineered human cells.
“It’s well known that nature has perfected very powerful information processing with only a small number of parts, but deconvoluting precisely how this works is virtually impossible in human cells due to their complexity,” adds Ellis, who was not part of the research. “By recreating the way human cells process information at the DNA level, but in a simple yeast cell model with synthetic parts, they have been able to recreate complex signaling from first principles. This is an excellent example of how thinking like an engineer can unlock a new way to answer major biology questions.”
###
Media Contact
Mike Seele
[email protected]
617-353-9766
Related Journal Article
http://dx.




