The technology for genome installation in model organisms
Credit: Rikkyo University
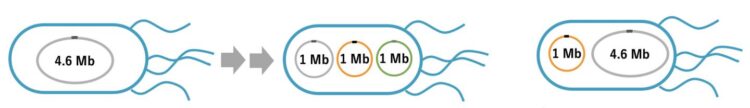
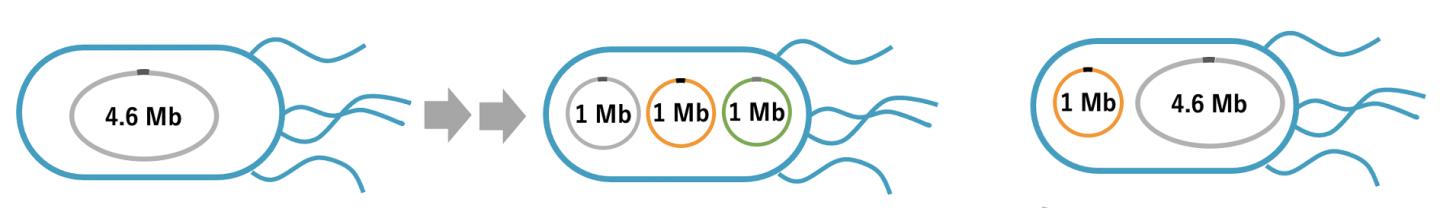
The issue of concern was that the Escherichia coli (E. coli) genome, consisting of 4.6 million base pairs of a single circular DNA, is too large to manipulate following the extraction and transfer to other bacteria.
In the present study, a group of Rikkyo University researchers led by Assistant Professor Takahito Mukai and Professor Masayuki Su’etsugu has succeeded in splitting the E.coli genome into tripartite-genome of 1 million base pairs per genome (split-genome) using the smallest E. coli genome strain established so far. In addition, they successfully extracted the split-genome from bacteria and installed it in other E. coli.
It is a major breakthrough that E. coli could stably proliferate even after the bacterial genome was split into tripartite-genome. Going forward, it is imperative to clarify how the replication and distribution of the tripartite-genome are controlled. Also, this research group has been developing the technology for synthesizing gigantic DNA without using cells (cell-free) and reported a cell-free technique for amplifying 1 million base pairs of circular DNA. In the future, the installation of cell-free synthesized split genomes in E. coli is expected to lead to the creation of artificial E. coli with designed valuable functions, such as material production.
This achievement is expected to lead to the clarification of the mechanism of genome replication/segregation and also to the application of tools in synthetic biology to convert the genome, the blueprint of life, so that we can create functionally designed life. The results in the present study have been published in the online version as a breakthrough paper in Nucleic Acids Research on April 28, 2021.
###
The research was supported by JST’s Strategic Basic Research Program, Core Research for Evolutional Science and Technology (CREST).
Media Contact
Masayuki Su’etsugu
[email protected]
Original Source
https:/
Related Journal Article
http://dx.