Meta-DNA structures transform the DNA nanotechnology world
Credit: Hao Yan
A team* of scientists from ASU and Shanghai Jiao Tong University (SJTU) led by Hao Yan, ASU’s Milton Glick Professor in the School of Molecular Sciences, and director of the ASU Biodesign Institute’s Center for Molecular Design and Biomimetics, has just announced the creation of a new type of meta-DNA structures that will open up the fields of optoelectronics (including information storage and encryption) as well as synthetic biology.
This research was published today in Nature Chemistry – indeed the meta-DNA self-assembly concept may totally transform the microscopic world of structural DNA nanotechnology.
It is common knowledge that the predictable nature of Watson-Crick base-pairing and the structural features of DNA have allowed DNA to be used as a versatile building block to engineer sophisticated nanoscale structures and devices.
“A milestone in DNA technology was certainly the invention of DNA origami, where a long single-stranded DNA (ssDNA) is folded into designated shapes with the help of hundreds of short DNA staple strands,” explained Yan. “However it has been challenging to assemble larger (micron to millimeter) sized DNA architectures which up until recently has limited the use of DNA origami.” The new micron sized structures are on the order of the width of a human hair which is 1000 times larger than the original DNA nanostructures.
Ever since gracing the cover of Science Magazine in 2011 with their elegant DNA origami nanostructures, Yan and collaborators have been working tirelessly, capitalizing on inspiration from nature, seeking to solve complex human problems.
“In this current research we developed a versatile “meta-DNA” (M-DNA) strategy that allowed various sub-micrometer to micrometer sized DNA structures to self-assemble in a manner similar to how simple short DNA strands self-assemble at the nanoscale level,” said Yan.
The group demonstrated that a 6-helix bundle DNA origami nanostructure in the sub-micrometer scale (meta-DNA) could be used as a magnified analogue of single-stranded DNA (ssDNA), and that two meta-DNAs containing complementary “meta-base pairs” could form double helices with programmed handedness and helical pitches.
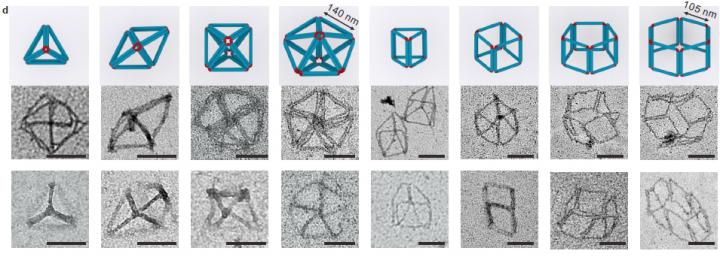
Using meta-DNA building blocks they have constructed a series of sub-micrometer to micrometer scale DNA architectures, including meta-multi-arm junctions, 3D polyhedrons, and various 2D/3D lattices. They also demonstrated a hierarchical strand-displacement reaction on meta-DNA to transfer the dynamic features of DNA to the meta-DNA.
With the help of assistant professor Petr Sulc (SMS) they used a coarse-grained computational model of the DNA to simulate the double-stranded M-DNA structure and to understand the different yields of left-handed and right-handed structures that were obtained.
Further, by just changing the local flexibility of the individual M-DNA and their interactions, they were able to build a series of sub-micrometer or micron-scale DNA structures from 1D to 3D with a wide variety of geometric shapes, including meta-junctions, meta-double crossover tiles (M-DX), tetrahedrons, octahedrons, prisms, and six types of closely packed lattices.
In the future, more complicated circuits, molecular motors, and nanodevices could be rationally designed using M-DNA and used in applications related to biosensing and molecular computation. This research will make the creation of dynamic micron-scale DNA structures, that are reconfigurable upon stimulation, significantly more feasible.
The authors anticipate that the introduction of this M-DNA strategy will transform DNA nanotechnology from the nanometer to the microscopic scale. This will create a range of complex static and dynamic structures in the sub-micrometer and micron-scale that will enable many new applications.
For example, these structures may be used as a scaffold for patterning complex functional components that are larger and more complex than previously thought possible. This discovery may also lead to more sophisticated and complex behaviors that mimic cell or cellular components with a combination of different M-DNA based hierarchical strand displacement reactions.
###
*ASU team members: Guangbao Yao (co first author , Fei Zhang (currently assistant professor at Rutgers University and co first author), Hao Liu, Erik Poppleton , Petr Šulc, Shuoxing Jiang, Lan Liu, Chen Gong, Yan Liu, Hao Yan (Co-corresponding author)
Team members from SJTU: Fei Wang (co first author), Tianhuan Peng, Xinxin Jing, Xiaoguo Liu, Lihua Wang, Chunhai Fan (Co-corresponding author)
Media Contact
Jenny Green
[email protected]




