Hepatocellular carcinoma (HCC), the predominant form of primary liver cancer, persists as a formidable global health challenge due to its high incidence and mortality rates. While traditionally linked to environmental risk factors such as hepatitis virus infections and chronic alcohol use, mounting evidence implicates epigenetic dysregulation as a pivotal mechanism driving hepatocarcinogenesis. Unlike genetic mutations that alter DNA sequences, epigenetic modifications reconfigure gene expression through reversible chemical changes, offering unique insights into tumor biology and promising avenues for therapeutic intervention.
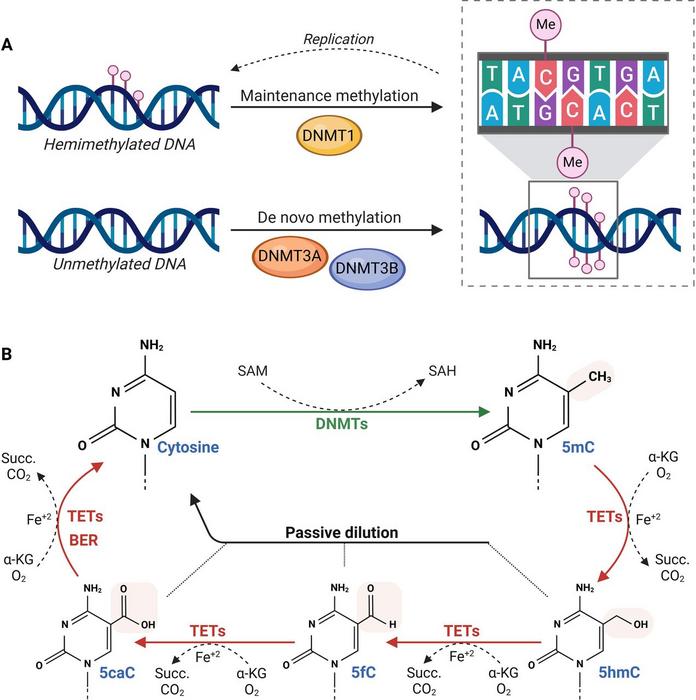
Central to epigenetic control is DNA methylation, a biochemical process involving the addition of methyl groups to cytosines in CpG dinucleotides, predominantly within gene promoter regions. This modification serves as a gatekeeper of gene expression, silencing or activating genes crucial for cellular homeostasis. In HCC, aberrant hypermethylation frequently targets tumor suppressor genes—such as CDKN2A, RASSF1A, and SOCS1—effectively silencing their protective functions. Such epigenetic repression disrupts canonical regulatory pathways governing cell cycle arrest and apoptosis, facilitating uncontrolled proliferation and tumor progression.
Conversely, global DNA hypomethylation represents another hallmark of HCC epigenetics, resulting in genomic instability and reactivation of normally silenced transposable elements and oncogenes. This dichotomous pattern—simultaneous hypermethylation of tumor suppressors and hypomethylation of oncogenic regions—vividly illustrates the complex remodeling of the cancer epigenome. Researchers have underscored the temporal dynamics of DNA methylation abnormalities, noting their emergence even at early pathological stages, thus positioning them as potential biomarkers for timely diagnosis.
.adsslot_dSxwpYUahf{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_dSxwpYUahf{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_dSxwpYUahf{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Beyond DNA methylation, chromatin architecture undergoes profound alterations through histone post-translational modifications. Histones, the core proteins around which DNA is wrapped, are subjected to a variety of chemical changes—including acetylation, methylation, phosphorylation, and ubiquitination—that dictate chromatin compaction and accessibility to transcriptional machinery. In hepatic malignancies, dysregulation of histone acetylation is especially prominent, with overexpressed histone deacetylases (HDACs) fostering chromatin condensation and gene silencing. This epigenetic silencing mechanism extends to tumor suppressors, further exacerbating oncogenic shift.
Histone methylation adds an additional layer of control, where the balance of activating and repressive marks guides gene transcription outcomes. Overexpression of histone methyltransferases such as EZH2 in HCC exemplifies this phenomenon, whereby tri-methylation of histone H3 lysine 27 leads to stable repression of tumor suppressor loci. Collectively, these histone modifications provide a versatile regulatory network that cancer cells exploit to regulate gene expression patterns favoring growth and metastasis.
Intriguingly, noncoding RNAs (ncRNAs) have emerged as master regulators within this epigenetic landscape. These molecules, which do not encode proteins, orchestrate a diverse array of gene regulatory processes. MicroRNAs (miRNAs) have garnered particular attention in HCC due to their capacity to modulate critical signaling cascades like Wnt/β-catenin, PI3K/Akt, and TGF-β pathways, impacting tumor cell proliferation and survival. Dysregulation of miRNAs such as miR-122, miR-221, and miR-21 is well documented in hepatic tumors, underscoring their oncogenic or tumor suppressive roles.
Long noncoding RNAs (lncRNAs), with their extensive sequences and structural complexity, engage in multifaceted interactions. Overexpressed lncRNAs including HULC, MALAT1, and HOTAIR not only function as molecular sponges sequestering tumor-suppressive miRNAs but also interface directly with chromatin modifiers, influencing epigenetic state and transcriptional programs. Circular RNAs (circRNAs) add further intricacy by acting as miRNA sponges and modulating gene networks involved in cell cycle regulation and metastasis. The interplay among ncRNAs and chromatin dynamics magnifies the oncogenic epigenetic rewiring characteristic of HCC.
This interconnectivity among epigenetic modifications engenders a complex regulatory circuit, wherein ncRNAs can influence DNA methylation and histone modification patterns, while histone modifiers reciprocally regulate the expression of ncRNA genes. This epigenetic crosstalk is thought to underlie the heterogeneity and aggressive clinical course of HCC, spotlighting these molecular players as attractive candidates for biomarker development and therapeutic targeting.
Excitingly, translational advances have begun to harness these epigenetic insights. Circulating methylated DNA panels show promise as noninvasive diagnostic tools for early HCC detection, potentially transforming patient outcomes through earlier interventions. Similarly, profiling circulating ncRNAs—including miRNAs and lncRNAs—in plasma relates closely to tumor burden and progression, offering prognostic utility.
Therapeutically, inhibitors of DNA methyltransferases (DNMTs) such as 5-azacytidine and decitabine have demonstrated efficacy in reactivating silenced tumor suppressor genes, attenuating tumor growth. Histone deacetylase inhibitors (HDACi), including vorinostat and belinostat, have yielded encouraging results by reversing aberrant chromatin compaction and sensitizing tumors to chemotherapy and immunotherapy. Furthermore, targeted modulation of ncRNAs using antisense oligonucleotides and miRNA mimics represents a cutting-edge approach to restore balanced gene regulation.
Despite these advances, significant challenges impede the clinical translation of epigenetic therapies. The intrinsic plasticity of epigenetic states enables tumor cells to evade inhibition through adaptive mechanisms, underscoring the need for combinatorial regimens and longitudinal monitoring. Moreover, most therapeutic candidates remain confined to early-phase studies, highlighting the urgent requirement for well-designed, large-scale clinical trials to validate efficacy and safety.
Future perspectives emphasize the integration of epigenomic data with complementary multi-omic platforms such as transcriptomics and proteomics, ushering in an era of precision hepatology. Such comprehensive profiles will refine molecular classifications, predict treatment responses, and reveal novel vulnerabilities in HCC. Concurrently, innovations in delivery methods, including nanoparticle-mediated systems and liver-targeted compounds, aim to enhance the specificity and minimize off-target effects of epigenetic drugs.
In summary, the evolving landscape of epigenetics in hepatocellular carcinoma provides profound mechanistic insights and unveils innovative possibilities for diagnosis, prognosis, and therapy. DNA methylation, histone modifications, and noncoding RNAs comprise a tightly interwoven regulatory network that orchestrates the oncogenic transformation of hepatocytes. As our understanding deepens, translating these molecular intricacies into clinical practice is poised to revolutionize HCC management, offering hope for improved patient survival and quality of life.
Subject of Research: Epigenetic mechanisms driving hepatocellular carcinoma development and progression
Article Title: Epigenetic mechanisms involved in hepatocellular carcinoma development and progression
News Publication Date: Not explicitly stated (article published in 2025)
Web References: http://dx.doi.org/10.1136/egastro-2025-100186
References: Bueloni B, Garcia Fernandez de Barrena M, Avila MA, et al. Epigenetic mechanisms involved in hepatocellular carcinoma development and progression. eGastroenterology 2025;3:e100186. doi:10.1136/egastro-2025-100186
Image Credits: Barbara Bueloni, Maite Garcia Fernandez de Barrena, Matias Antonio Avila, Juan Bayo, Guillermo Mazzolini
Keywords: hepatocellular carcinoma, epigenetics, DNA methylation, histone modification, noncoding RNA, biomarkers, DNMT inhibitors, HDAC inhibitors, microRNA, long noncoding RNA, circular RNA, tumor suppressor silencing, oncogene activation
Tags: DNA methylation and cancerenvironmental risk factors for liver cancerepigenetic dysregulation in hepatocarcinogenesisepigenetic mechanisms in hepatocellular carcinomagene expression modifications in cancerglobal hypomethylation and genomic instabilityhepatocellular carcinoma prevalence and mortalityhypermethylation in liver cancerliver cancer biological pathwaysoncogene reactivation in HCCtherapeutic intervention in liver cancertumor suppressor gene silencing in HCC