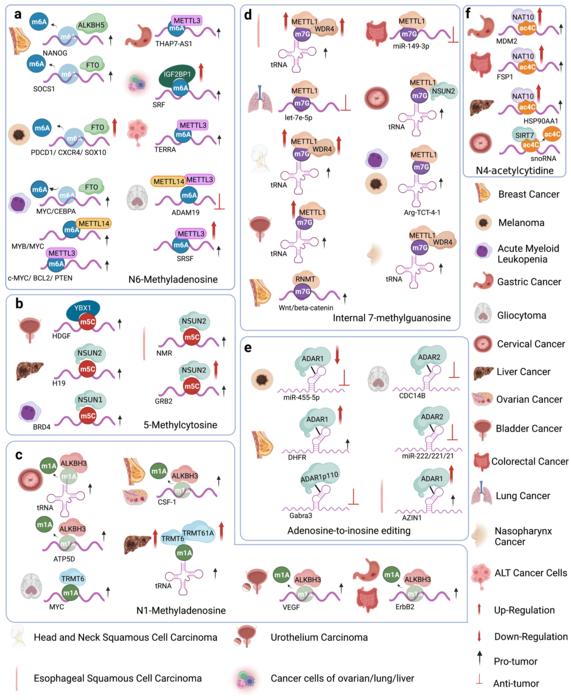
This review is designed by Dr. Junhong Han and written by his postdoctoral researcher Dr. Lei Qiu and PhD student Qian Jing (Research Laboratory of Tumor Epigenetics and Genomics, West China Hospital, Sichuan University). RNA modifications are dynamic and reversible chemical modifications on substrate RNA that regulate mRNA stability, translation, and localization. Currently identified RNA modifications are modulated by the “writing-erasing-reading” system, which consists of various RNA modifying enzymes functioning as “writers”, “erasers” and “readers”. In a complete process of RNA modification, “writer” installs RNA modifications on RNA substrates; “eraser” removes the chemical marks installed on RNA; “reader” recognizes the RNA modifications and guides the fate of target RNAs. Qiu and Jing reviewed the specific RNA modifiers and the detailed mechanisms of six common RNA modifications, including m6A, m5C, m1A, m7G, Ψ, and A-to-I editing. Among the six RNA modifications, m6A, m5C, m1A, and m7G belong to the RNA methylation category, whereas Ψ and A-to-I editing are RNA editing. They also mentioned a novel RNA modification, ac4C, which is installed by NAT10 and possibly removed by SIRT7.
Credit: Beijing Zhongke Journal Publising Co. Ltd.
This review is designed by Dr. Junhong Han and written by his postdoctoral researcher Dr. Lei Qiu and PhD student Qian Jing (Research Laboratory of Tumor Epigenetics and Genomics, West China Hospital, Sichuan University). RNA modifications are dynamic and reversible chemical modifications on substrate RNA that regulate mRNA stability, translation, and localization. Currently identified RNA modifications are modulated by the “writing-erasing-reading” system, which consists of various RNA modifying enzymes functioning as “writers”, “erasers” and “readers”. In a complete process of RNA modification, “writer” installs RNA modifications on RNA substrates; “eraser” removes the chemical marks installed on RNA; “reader” recognizes the RNA modifications and guides the fate of target RNAs. Qiu and Jing reviewed the specific RNA modifiers and the detailed mechanisms of six common RNA modifications, including m6A, m5C, m1A, m7G, Ψ, and A-to-I editing. Among the six RNA modifications, m6A, m5C, m1A, and m7G belong to the RNA methylation category, whereas Ψ and A-to-I editing are RNA editing. They also mentioned a novel RNA modification, ac4C, which is installed by NAT10 and possibly removed by SIRT7.
Based on the RNA modification regulatory system, Qiu and Jing comprehensively summarized the functional involvement of the seven RNA modifications in five human disease models, including cancer, neurological disorders, cardiovascular disease, metabolic disorders, as well as genetic and developmental diseases. In detail, they discussed the impact of dysregulation of each specific RNA modifier in different diseases. For example, the m6A “writer” METTL3/METTL14 showed both oncogenic and tumor suppressive roles depending on the target transcript and disease model. Genetic birth defects and developmental defects involve mutations of m5C “writers”, such as the Cri du chat syndrome (NSUN1) and the Dubowitz syndrome (NSUN2). m7G methyltransferase METTL1 promoted blood flow recovery in cardiovascular disease.
Importantly, they listed nearly all available promising small molecule inhibitors targeting various RNA modifiers and briefly explained their acting mechanisms. Generally, most of the inhibitors were developed against RNA methylation, especially those inhibitors specifically targeting m6A modifying enzymes such as METTL3 and FTO.
Based on a thorough understanding and summary of the mechanisms of RNA modifications, their roles in disease models, and their targeting small molecules, Qiu and Jing also provided novel insights regarding future directions of research in the RNA modification field. This comprehensive review is beneficial to the development of novel RNA-modification-targeted therapeutic strategies.
Journal
Molecular Biomedicine
DOI
10.1186/s43556-023-00139-x
Article Title
RNA modification: mechanisms and therapeutic targets
Article Publication Date
24-Aug-2023



