When we’re driving to a new destination, we often turn down the stereo as we follow the directions. What had been music suddenly sounds like noise, and it interferes with our focus.
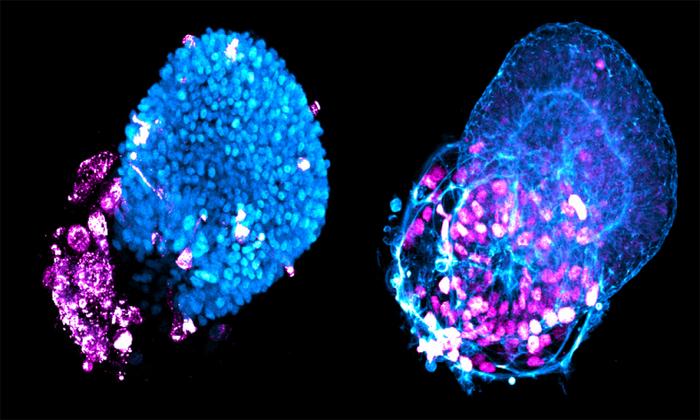
Credit: Laboratory of Synthetic Biology at The Rockefeller University
When we’re driving to a new destination, we often turn down the stereo as we follow the directions. What had been music suddenly sounds like noise, and it interferes with our focus.
Our understanding of how infectious diseases like COVID affect human lungs has been similarly confounded by noise. Data from patient lung tissues greatly varies from person to person, obscuring the basic mechanisms of how, exactly, SARS-Co-V2 first infects lung cells. It’s also an after-the-fact analysis—as if we’re trying to map the route the virus took three states back.
Turning down the noise of variability by studying genetically identical tissues from the first moment of infection could light up the route the pathogen takes. Which cells are infected, and when? What is the level of infection, and how does it differ depending on cell type? How does it change in different conditions?
And what if it were possible to track thousands of these infections at once? It might revolutionize our understanding of both infections and the drug treatments used to combat them.
That’s the hope for new advanced tech capable of growing mini organs on microchips. The labs of Rockefeller’s Ali Brivanlou and Charles M. Rice collaborated to refine a cell culture technology platform that grows genetically identical lung buds—the embryonic structures that give rise to our breathing organs—from human embryonic stem cells (hESCs). Their findings were recently published in Stem Cell Reports.
When placed on an array of microchips and carefully dosed with a custom cocktail of signaling molecules, the hESCs rapidly organize themselves into “micro lungs” that have full tissue complexity. These buds can be cultured by the thousands, allowing for an unprecedented high-throughput analysis of lung tissue infection without all the noisy variables.
The result is unlimited, fast, and scalable access to lung tissue that has the key hallmarks of human lung development and can be used to track lung infections and identify candidate therapeutics.
“These lungs are basically clones,” says Ali Brivanlou. “They have the exact same DNA signature. That way we don’t have to worry about one patient responding differently from another. Quantification allows us to keep the genetic information constant and measure the key variable—the virus.”
Building a better mini lung
Embryonic stem cells are the Ur-cells of the human body. They can infinitely divide to create more stem cells or to differentiate into any other tissue. Brivanlou’s Laboratory of Synthetic Biology has long explored their potential.
Brivanlou joined forces with Rockefeller colleague Charles M. Rice during the COVID pandemic: his lab had the microchip technology to grow lung buds, and Rice’s lab had the necessary biosafety clearance required to infect them with SARS-Co-V2 and study the outcome.
In 2021, first authors Edwin Rosado-Olivieri, a stem cell biologist in Brivanlou’s lab, and Brandon Razooky, then a postdoc in Rice’s Laboratory of Virology and Infectious Disease, began coaxing the cells to organize into more specialized forms. Stem cells don’t just organize on their own. They need a confined space—such as a microchip well—and stimuli to spark change. The stimuli come from four main signaling pathways that induce stem cells to differentiate into specific cell types.
After about two weeks, the group’s lung cells had formed identical buds whose molecular profiles closely matched those seen in the earliest stages of fetal lung development—including the formation of airways and alveoli, structures known to be damaged in many people with severe COVID.
Identifying a key culprit
Since then, they’ve used the platform to understand how SARS-Co-V2 infects different lung cells.
Alveoli are tiny sacs at the end of the lung branches that manage the gas exchange performed with every breath: oxygen in, carbon dioxide out. By studying cloned alveoli cells en masse, the researchers discovered that alveoli were more susceptible to SARS-Co-V-2 infection than airway cells, which are the guardians of the organ—the first defense against all inhaled threats. If the virus got past them, the alveoli were sitting ducks.
Another view of virus particles (blue) infecting alveolar and airway tissues (red).
They also hit upon a winning combination of signaling proteins for creating the most robust batches of lung buds—a mix of keratinocyte growth factor (KGF) and bone morphogenetic protein 4 (BMP4). Both contribute to cell differentiation and growth.
Interestingly, the BMP pathway has a downside. When they compared infected lung buds to postmortem tissue of COVID patients, they found that the BMP signaling pathway was induced in both and rendered the tissues more vulnerable to infection. Blocking the BMP pathway made the cells less vulnerable.
Beyond COVID
The researchers note that the platform can also be used to investigate the mechanisms of influenza, RSV, pulmonary diseases, and lung cancer, among other diseases. Moreover, it can be used to screen for new drugs to treat them.
And lungs are far from the only organ of interest. “The broader focus of our work is understanding cellular development to make synthetic organs and tissues that we can use to model diseases and find therapeutic mechanisms,” says Rosado-Olivieri. The liver, kidney, and pancreas are all likely next targets.
“The platform will also allow us to respond to the next pandemic with much more speed and precision,” Brivanlou adds. “We can quickly capitalize on this platform to make a virus visible and develop therapies much faster than we did for COVID. It can be used to screen for drugs, compounds, vaccines, monoclonal antibodies, and more directly in human tissue. This technology is ready to confront all kinds of threats that may hit us in the future.”
Journal
Stem Cell Reports
DOI
10.1016/j.stemcr.2023.03.015