Helper protein controls interactive process of ribosome formation
Credit: Nikolay/Charité.
Ribosome formation is viewed as a promising potential target for new antibacterial agents. Researchers from Charité – Universitätsmedizin Berlin have gained new insights into this multifaceted process. The formation of ribosomal components involves multiple helper proteins which, much like instruments in an orchestra, interact in a coordinated way. One of these helper proteins – protein ObgE – acts as the conductor, guiding the entire process. The research, which produced the first-ever image-based reconstruction of this process, has been published in Molecular Cell*.
Ribosomes are an essential component of all living cells. Frequently referred to as ‘molecular protein factories’, they translate genetic information into chains of linked-up amino acids which are otherwise known as proteins. The process of protein biosynthesis is the same in all cells, even in bacteria (including the widely known intestinal bacterium Escherichia coli). If this process cannot take place, the cell dies; single-celled organism (such as E. coli and other bacteria) cannot survive. Researchers are hoping to exploit this circumstance for the development of novel antibiotic agents. The need for these new drugs is not only the result of an increase in antibiotic resistance and the emergence and spread of new multidrug-resistant pathogens, but also because it has been a long time since a new class of antibiotic substances emerged. A new type of antibiotic might be designed to interfere with ribosome formation in a way that inhibits their assembly.
“It is a coincidence that we are currently in the middle of a viral pandemic. The next pandemic could easily be of bacterial origin because both bacterial antibiotic resistance and multidrug resistance are spreading rapidly, across species barriers”, explains the study’s last author, Prof. Dr. Christian Spahn, Director of Charité’s Institute of Medical Physics and Biophysics. He adds: “The long-term aim of our basic research is therefore to contribute to the development of new antibiotics.” Working with colleagues from the Max Delbrück Center for Molecular Medicine MDC) in Berlin and the University of Konstanz, the Charité researchers explored the early stages of ribosome formation to identify points in the process which might serve as targets for new antibacterial and antimicrobial drugs.
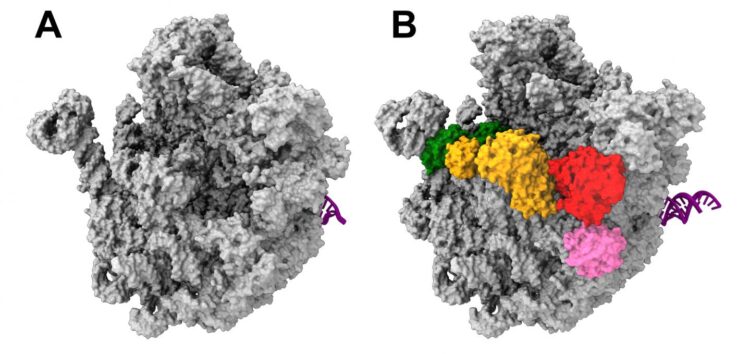
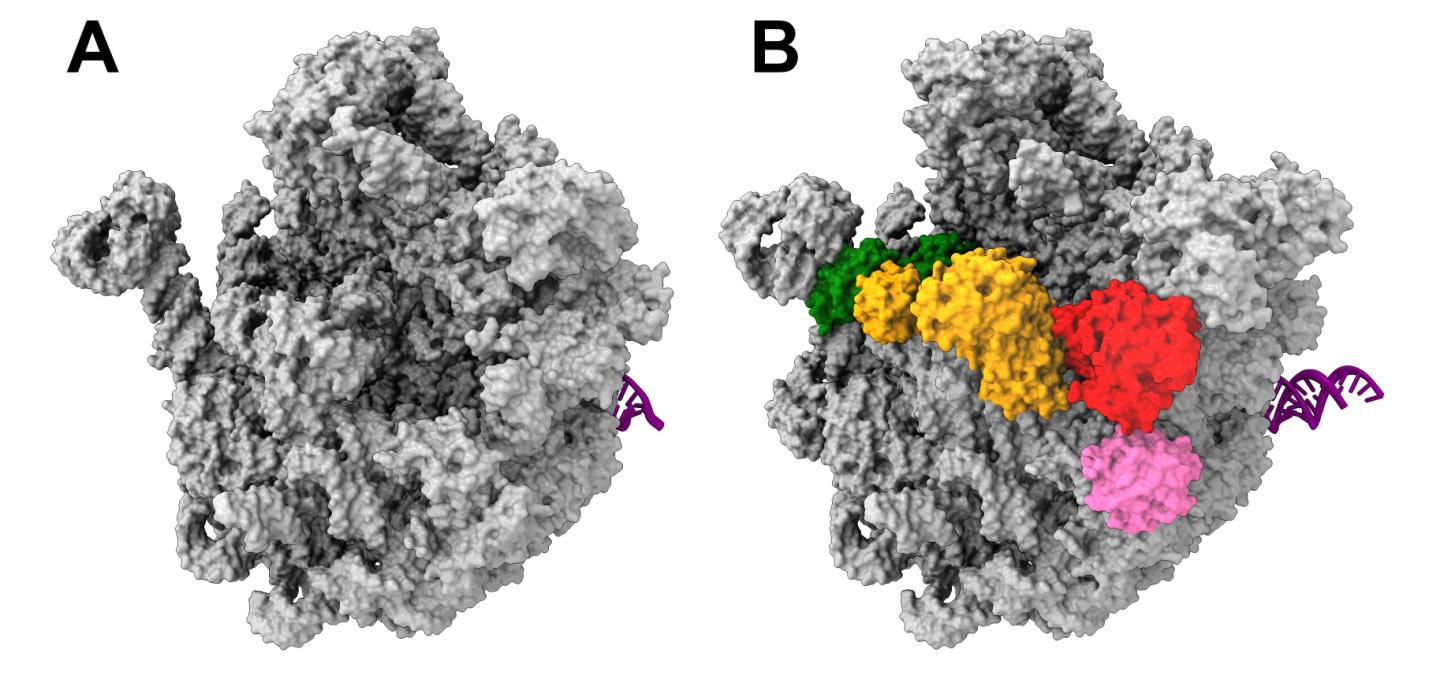
Ribosomes consist of two subunits: one larger subunit and one smaller one. As part of their latest endeavors, the team, led by Dr. Rainer Nikolay of Charité’s Institute for Medical Physics and Biophysics, focused on studying the nature and development of the larger ribosomal subunit in the bacterium E.coli. Hoping to identify a potential target for new antibiotics, the researchers wanted to isolate and visualize the precursor stages of this larger subunit. To do so, they wanted to use the subunit in its unadulterated form, i.e. as close to its natural condition as possible. For the first time, the researchers succeeded in not only isolating one such precursor from bacterial cells (in this case, E. coli), but also visualizing it using cryo-electron microscopy imaging at near-atomic resolution. “We now have a better understanding of the way in which the larger bacterial ribosomal subunit develops at the molecular level, although our understanding remains far from complete,” says first author Dr. Nikolay.
The research team chose a minimally invasive protocol in order to minimize the degree to which the bacterial cell would need to be manipulated. One of the key agents in the process of ribosome formation, the protein ObgE, was marked using what is known as a ‘Strep tag’. This step involves a ‘gene knock-in’ procedure – the insertion of genetic information into the bacterial genome. A bacterium thus treated will produce only marked ObgE. After minor processing of the cell, this ObgE can then be visualized using an electron microscope. Strep tagging enabled the researchers to study the entire complex for the first time. This is because the helper protein ObgE effectively carries the precursor of the larger ribosomal subunit on its back. The results came as a surprise, as Dr Nikolay explains: “We found that this precursor is covered in multiple helper proteins, which either interact or directly communicate with one another. The ObgE protein takes on a key role in this process, effectively directing and coordinating it.” This could constitute a target for new drugs, which might stop bacterial growth by inhibiting the assembly of functional ribosomes.
The team want to use similar strategies to gain further insights into the development of bacterial ribosomal subunits and enhance their understanding of the relevant biological processes at the molecular level. Previous research, conducted at Charité and the Max Planck Institute for Molecular Genetics, had produced valuable information on the fundamental structure of ribosomes and the various steps of the maturation process which these cellular protein factories must undergo. While all of these earlier insights were based on in vitro studies, the researchers knew that the formation of the large chromosomal subunit could only be observed in a living cell. The latest step in their endeavors has therefore been a crucial one: in order to identify new cellular drug targets, it is necessary to understand how the process of ribosome formation seen in bacteria differs from that in human cells. “We have managed to make some headway in that respect,” says Dr. Nikolay. “We were able to reveal the existence of both conserved and divergent evolutionary features between prokaryotes – such as bacteria – and eukaryotes – organisms whose genetic information is contained inside a cell nucleus.” These findings are important if we are to target features specific to bacteria while also protecting human cells against unwanted side effects.
###
*Nikolay R et al. Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization. Mol Cell. 2021 Feb 26. Doi: 10.1016/j.molcel.2021.02.006
Media Contact
Dr. Christian Spahn
[email protected]
Original Source
https:/
Related Journal Article
http://dx.