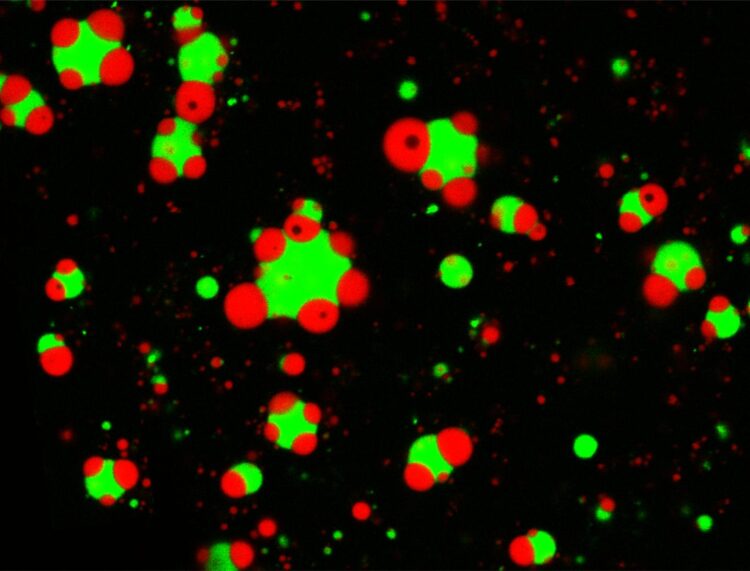
A study in Nature Communications outlines physical rules regulating the architecture of these liquid organelles
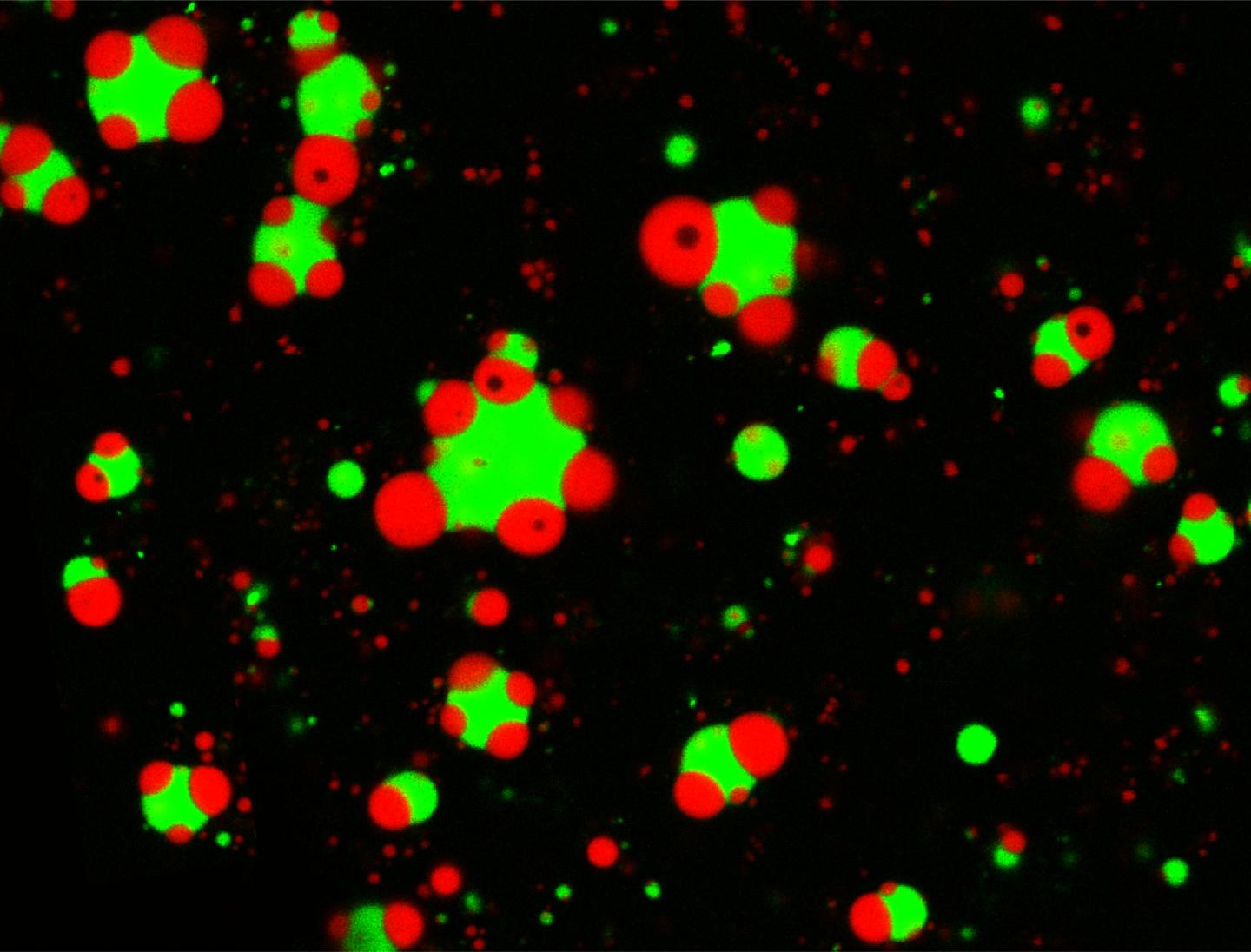
Credit: Taranpreet Kaur
BUFFALO, N.Y. — In cells, numerous important biochemical functions take place within spherical chambers made from proteins and RNA.
These compartments are akin to specialized rooms inside a house, but their architecture is radically different: They don’t have walls. Instead, they take the form of liquid droplets that don’t have a membrane, forming spontaneously, similar to oil droplets in water. Sometimes, the droplets are found alone. Other times, one droplet can be found nested inside of another. And these varying assemblies can regulate the functions the droplets perform.
A study published on Feb. 8 in Nature Communications explores how these compartments, also known as membraneless organelles (MLOs) or biomolecular condensates, form and organize themselves. The research lays out physical rules controlling the arrangement of various types of synthetic MLOs created using just three kinds of building materials: RNA and two different proteins, a prion-like polypeptide (PLP) and an arginine-rich polypeptide (RRP).
The project brought together a team from the University at Buffalo and Iowa State University.
“Different condensates can coexist inside the cells,” says first author Taranpreet Kaur, a PhD student in physics in the UB College of Arts and Sciences. “They can be detached, attached to another condensate, or completely embedded within one another. So how is the cell controlling this? We found two different mechanisms that allowed us to control the architecture of synthetic membraneless organelles formed inside a test tube. First, the amount of RNA in the mixture helps to regulate the morphology of the organelles. The other factor is the amino acid sequence of the proteins involved.”
“These two factors impact how sticky the surfaces of the condensates are, changing how they interact with other droplets,” says Priya Banerjee, PhD, UB assistant professor of physics, and one of two senior authors of the paper. “In all, we have shown using a simple system of three components that we can create different kinds of organelles and control their arrangement in a predictive manner. We suspect that such mechanisms may be employed by cells to arrange different MLOs for optimizing their functional output.”
Davit Potoyan, PhD, assistant professor of chemistry at Iowa State University, is the study’s other senior author.
Addressing questions in cell biology
The experiments were done on model systems made from RNA and proteins floating in a buffer solution. But the next step in the research — already underway — is to conduct similar studies inside a living cell.
“Going back to our motivations in researching MLOs, the big questions that started the field were questions in cell biology: How do cells organize their internal space?” Banerjee says. “The principles we uncover here contribute to the knowledge base that will improve understanding in this area.”
Research on MLOs could lead to advancements in fields such as synthetic cell research or new materials for drug delivery.
“We are in the process of learning the biomolecular grammar that may be a universal language used by cells for taming their inner cellular complexity. We hope one day to utilize this knowledge to engineer artificial protocells with custom-designed functionalities inspired by nature,” Potoyan says.
###
In addition to Banerjee, Potoyan and Kaur, co-authors of the study included Iowa State University chemistry postdoctoral researcher Muralikrishna Raju; UB physics PhD student Ibraheem Alshareedah; and UB physics postdoctoral researcher Richoo Davis.
The study was supported by the National Institute of General Medical Sciences, part of the U.S. National Institutes of Health, and the U.S. National Science Foundation (NSF). The team also received assistance from two NSF-funded resources: The UB North Campus Confocal Imaging Facility, and the Extreme Science and Engineering Discovery Environment.
Media Contact
Cory Nealon
[email protected]
Original Source
http://www.
Related Journal Article
http://dx.