University of Illinois biophysicists use Frontera supercomputer to study the formation and behavior of biological condensates
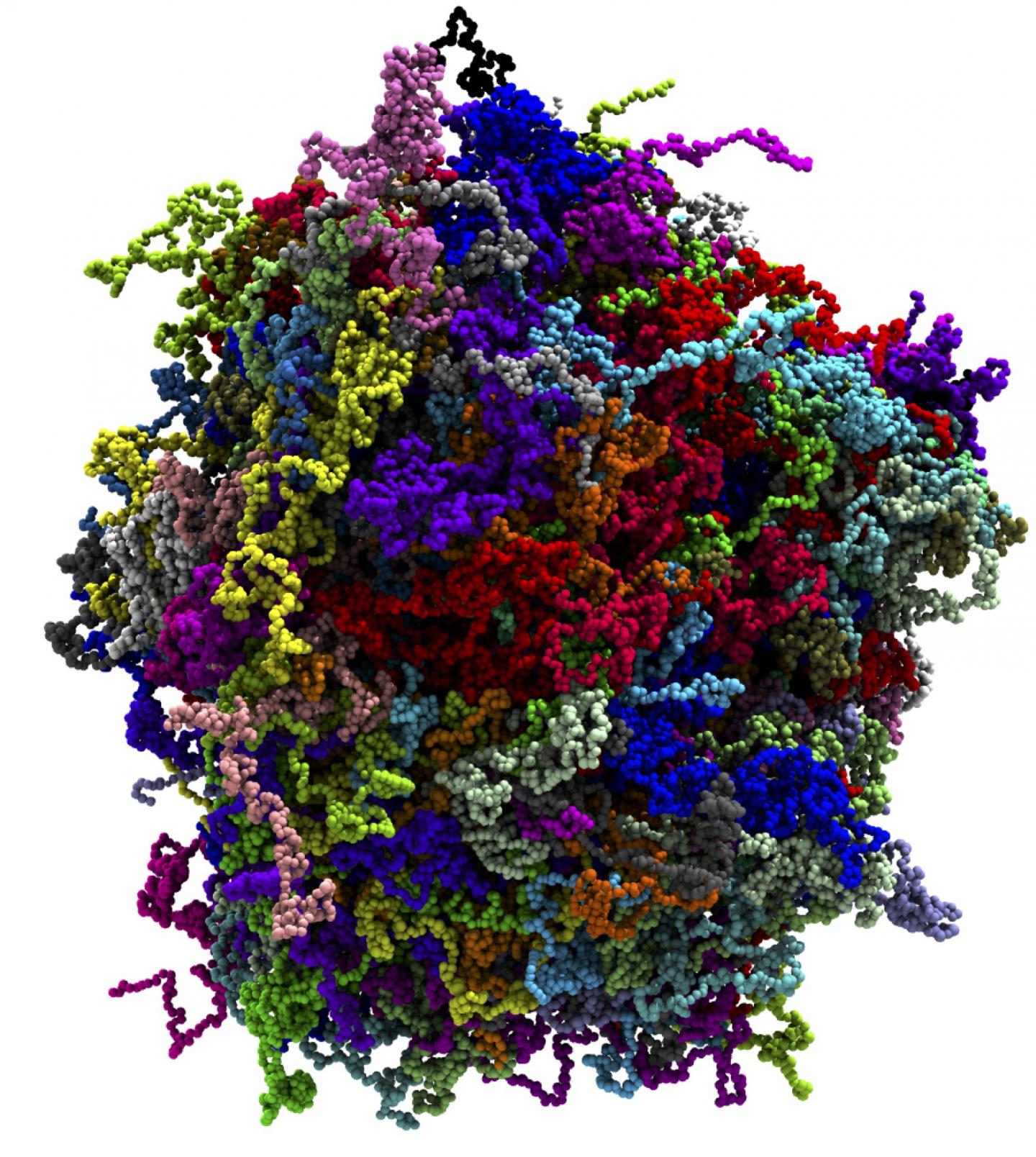
Credit: Han-Yi Chou, University of Illinois, Urbana-Champaign
The world inside the human cell grew a bit more interesting in recent years as the role of a new biological structure became clearer.
It was long believed that most important operations in the cell occur within organelles. “They’re there to do certain functions. For instance, mitochondria generate the energy that everything runs on,” explained Aleksei Aksimentiev, a professor of physics at the University of Illinois at Urbana-Champaign. “What is common to all of them is that they’re surrounded by a lipid membrane. What people recently discovered is there are organelles that don’t have lipid bilayers. They assemble spontaneously in the form of droplets. And those organelles have particular functions.”
In recent years, with improved imaging capabilities, the roles, occurrence, and behavior of these membrane-less organelles have become clearer. In 2017 they were given a name: biological condensates. They are thought to play a role in DNA repair and aging, and researchers believe a number of neurological diseases are related to the condensate not working properly, including Amyotrophic lateral sclerosis, or ALS, where nerve cells break down, leading to loss of muscular function.
“Let’s say you have DNA and it suddenly has a break. It’s usually a really bad thing, because it cannot replicate, but there’s a machinery that will come and repair it,” he explained. “A bubble of condensate forms that miraculously attracts only the molecules that are required to repair the DNA. There are all kinds of different condensates and they all recruit the right molecules somehow.”
How do these membrane-less organelles spontaneously form? And how do they recruit other molecules to help them?
The physics of this process appears similar to phase separation, like how oil and water spontaneously form droplets in the right conditions, but with some differences. In normal phase separation, temperature usually motivates the separation. In biology, it is a change in concentrations.
“We don’t know exactly how it works,” Aksimentiev said. “I’m specifically interested in how this recruitment happens, and how molecules recognize other molecules.”
Aksimentiev is using the Frontera supercomputer at the Texas Advanced Computing Center (TACC), one of the fastest in the world, to better understand this process. Over the last decade, he and others developed the tools and methods to explore the behavior of biological systems at the atomic level using molecular dynamics simulations.
Aksimentiev is able to simulate biological systems with millions of interacting atoms in a realistic environment for microseconds or even milliseconds — the timescales at which biological systems operate. Today’s supercomputers allow larger, faster simulations, and permit scientists to ask and answer new questions.
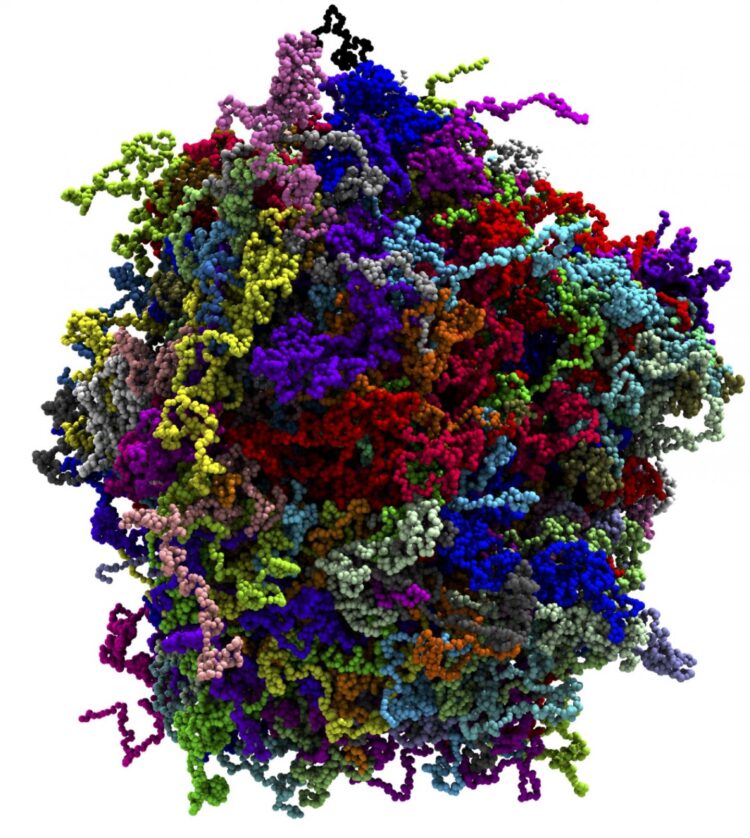
Even by the standards of the field, biological condensates are challenging to study computationally. Unlike other ordered systems like proteins with known rigid structures, or disordered systems like water, biological condensates are what’s known as ‘partially disordered’ — a particularly hard type of structure to simulate.
Writing in the Journal of Physical Chemistry Letters in May 2020, Aksimentiev and graduate student Han-Yi Chou described coarse-grained molecular dynamics simulations on Frontera that charted the phase diagram (a graphical representation of the physical states of a substance under different conditions of temperature and pressure) of one particular biomolecular condensate — fused in sarcoma (FUS). A nuclear DNA/RNA binding protein, FUS regulates different steps of gene expression, including transcription, splicing and mRNA transport. The research was supported by grants from the National Science Foundation and the National Institutes of Health.
The researchers showed that a particle-based molecular dynamics model can reproduce known phase separation properties of a FUS condensate, including its critical concentration and susceptibility to mutations.
They also showed that they could use chain collapse theory to determine the thermodynamic properties of the condensate and to link them to changes in the shape of individual condensate molecules.
The behavior of a biological condensate, with all its complex inter- and intramolecular interactions, can be described by a polymer physics model, they found. This makes computer modeling a useful tool for uncovering the behavior of these still-mysterious cellular actors.
Aksimentiev’s research sets the stage for future studies that will elucidate the molecular mechanisms driving the formation of droplets in more complex biological condensates, like those that repair RNA. The work is one step on a long path to fully elucidate the mystery of biological condensates in cells — another trick of nature slowly uncovered.
###
Media Contact
Aaron Dubrow
[email protected]
Original Source
https:/
Related Journal Article
http://dx.