Credit: © 2020 Simply Science Illustration
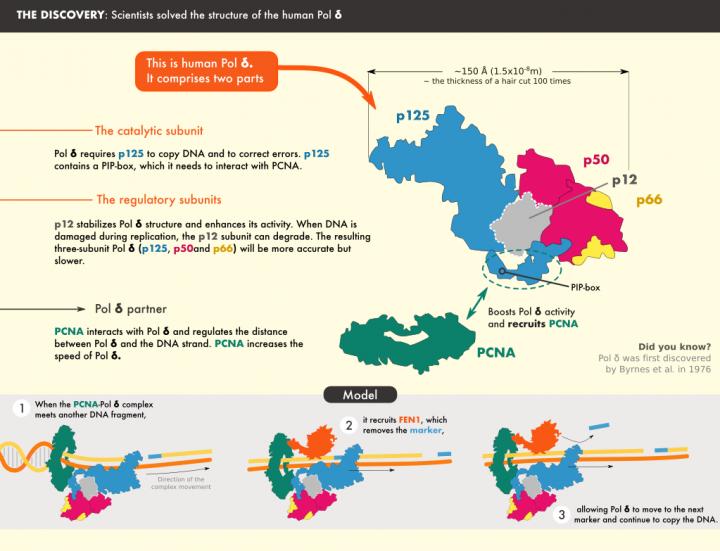
The molecular machinery responsible for replicating human DNA has come into sharper focus and deepened understanding of this fundamental biological process and how it can malfunction. An international team led by researchers at KAUST has used cryo-electron micrography to study the structure of the human Pol δ-DNA-PCNA complex.
During replication, the two strands in the DNA helix are unwound, and each serves as the template for a new copy. Pol δ is one of the enzymes responsible for copying one of the strands. It is tethered to the DNA by a molecule known as PCNA, forming a Pol δ-DNA-PCNA complex at the heart of replication.
The team found that the catalytic core of Pol δ–the part that carries out the synthesis reaction–sits on top of PCNA and that its regulatory subunits project to the side. The DNA is threaded through a central channel in PCNA, enabling the molecule to stabilize DNA exiting the reaction area. Further analysis revealed locations in PCNA that are required for the interaction, and results showed that mutating those sites severely reduced the activity of Pol δ.
Muhammad Tehseen, one of the lead authors of the study, explains that “Professor Hamdan’s lab has been systematically reconstituting and studying the activities of these proteins by single-molecule imaging, recording molecular movies of these reactions.”
Tehseen purified the molecular complex so that the team could create higher resolution static images. “Combining the dynamic information from single-molecule imaging with static images from cryoEM is like bringing an image to life,” he explains.
The structure also clarified the role of some of the Pol δ subunits, shedding light on how it interacts with PCNA and DNA. For example, Pol δ interacts with only one monomer of PCNA, and sits in a manner that exposes the two other monomers, freeing them to interact with other proteins that act with Pol δ.
One such protein is FEN1, which processes the short fragment strands of DNA, after they are synthesized by Pol δ, so that they can be stitched together to form a continuous mature strand.” The researchers also studied the structure of the Pol δ-DNA-PCNA-FEN1 complex. The interactions they discovered support a “toolbelt” model in which PCNA acts as a platform to which different processing enzymes bind, similar to a toolbelt with an array of tools.
Working out these structures and interactions enables researchers to better understand the dynamics of DNA replication and how it goes wrong, such as mutations in Pol δ linked with tumors.
###
Media Contact
Carolyn Unck
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




