Credit: Pavel Odinev / Skoltech
Researchers from the Severinov Laboratory at Skoltech, along with their colleagues from Switzerland and Israel, have investigated a poorly studied bacterial BREX defense mechanism to show that it can be “turned off” by a multipurpose viral protein that successfully impersonates DNA. The paper was published in the journal Nucleic Acids Research.
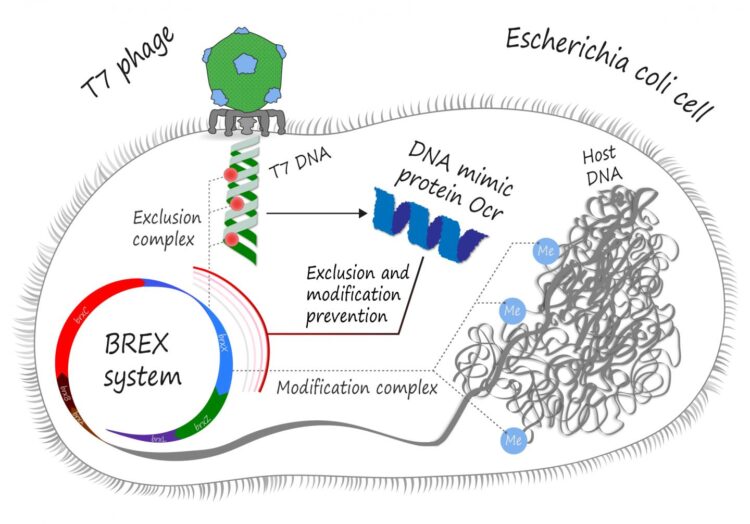
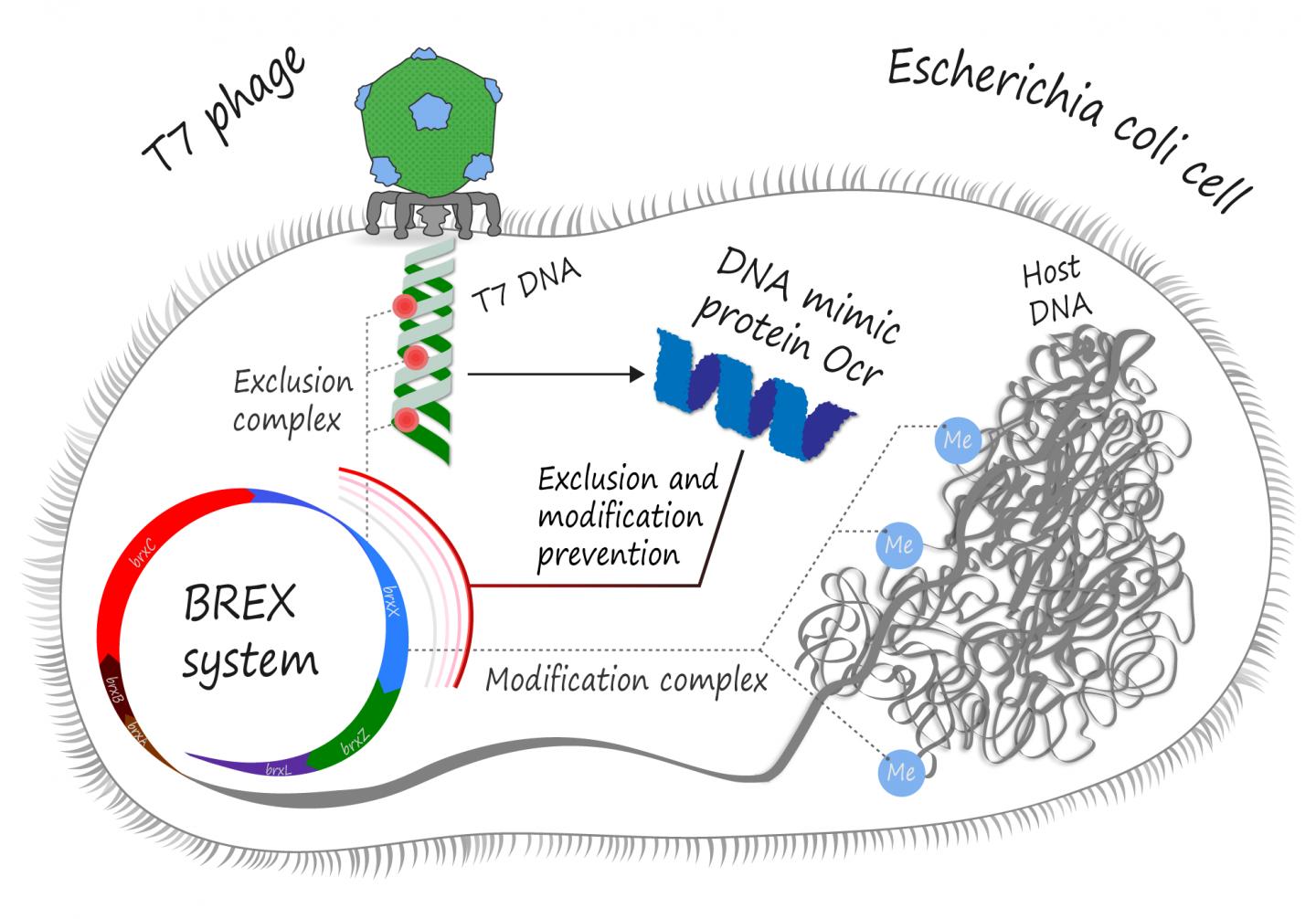
In the never-ending war between bacteria and viruses that infect them, it is extremely important to know where your DNA is. To protect themselves from hostile invasion, bacteria have learned to “mark” their own genetic material by methylating it at specific sites in the genome. All “unmarked” DNA, such as that of a bacteriophage, is then recognized, cleaved and degraded by an army of endonucleases. These are called restriction modification (RM) systems. The phages, in turn, have learned to evade these RM systems by using DNA mimic proteins. A protein that chemically “looks” like DNA to the bacterial restriction complex can bind it and prevent from ever getting to actual phage DNA.
Skoltech PhD student Artem Isaev and his colleagues from Tel Aviv University and Philip Morris International R&D have shown that Ocr, a DNA mimic protein of the well-studied T7 phage, is in fact a multipurpose tool. Besides inhibition of RM type I systems, it can also protect the phage from BREX (for BacteRiophage EX?lusion), another set of bacterial defense mechanisms utilized by, among others, Escherichia coli, which T7 commonly infects.
“Five years ago, we knew about restriction modification, CRISPR and Toxin-Antitoxin abortive infection systems, but recently bioinformatics has shown us that it is just a small proportion of the real diversity of defensive strategies employed by bacteria to cope with phage infections. BREX was the first in a row of novel phage defense systems: they are found in around 1 in 10 of all microorganisms, and in bacteria they are even more widespread than CRISPR. Yet we still don’t know the function of five out of six BREX genes and how they are working together to provide protection,” Isaev says.
It is yet unknown whether BREX simply destroys the incoming phage DNA or somehow inhibits its replication, but almost all BREX mechanisms employ a BrxX methyltransferase, an enzyme in charge of “marking” bacterial DNA for self-recognition. The Ocr protein apparently binds to this methyltransferase and thus prevents the BREX system from both methylation of the host DNA and attacking the non-methylated phage genome.
“We have shown that Ocr interacts with methyltransferase and inhibits host DNA methylation. In theory, this can lead to an autoimmune response: as host DNA is no longer “marked” as such, BREX exclusion complexes should attack it. Yet, we do not see self-toxicity after expression of Ocr, which indicates that Ocr inhibits these exclusion complexes as well, and thus BREX methyltransferase should be involved at the active stage of defense. Ocr is already known as an inhibitor of type I RM systems, and these systems also require methytransferase for restriction complexes. There are other similarities between BREX and RM systems, and we hope that they would help us understand how BREX functions,” Isaev explains.
Other DNA mimic proteins do not seem to overcome the BREX defense, so researchers intend to further investigate how exactly Ocr does its job. As bacterial defense systems mostly deal with DNA recognition and manipulation, they can become powerful tools for molecular biology and medicine. Molecular cloning is possible thanks to the discovery and description of RM systems, and CRISPR has brought about the age of genome editing. Moreover, studying the arsenals of bacteria and phages may prove useful in “recruiting” the viruses as novel antimicrobial agents in the fight against antibiotic-resistant bacteria.
“Bacteria have been combatting phages for more than a billion years, and this constant “arms race” is one of the major evolutionary forces in the microworld. Both sides have developed an enormous arsenal of strategies to fight each other, and a great diversity of molecular machines has been invented in the process. For me personally, it’s just fascinating to study what else is hidden in the genome and what novel mechanism we can discover in the process,” Isaev concludes.
###
Media Contact
Alina Chernova
[email protected]
Related Journal Article
http://dx.