Credit: Tokyo Metropolitan Institute of Medical Science
The proteasome is a major proteolytic machine that regulates cellular proteostasis through selective degradation of ubiquitylated proteins. As the maintenance of protein homeostasis is essential to human health, malfunctions of the ubiquitin-proteasome system (UPS) causes various diseases such as cancers, inflammation, and neurodegeneration. However, we do not yet know the overall principles behind UPS.
In this study, the research team discovered a novel mode of protein degradation by the UPS under a stress condition.
“Around 2013, we discovered that proteasomes form nuclear foci in response to hyperosmotic stimuli. It was very impressive because the uniformly distributed proteasomes form foci in just a few seconds, but it took time to understand what this phenomenon means until we know liquid-liquid phase separation (LLPS). LLPS is a rapid, reversible, and wide-spread compartmentalization mechanism in cells. The proteasome foci actually exhibit liquid-like behavior and a series of experiments revealed the LLPS of proteasomes is for degradation of ubiquitylated proteins.” said Yasushi Saeki, PhD, a lead author of the study.
The research results were published in Nature on February 5, 2020.
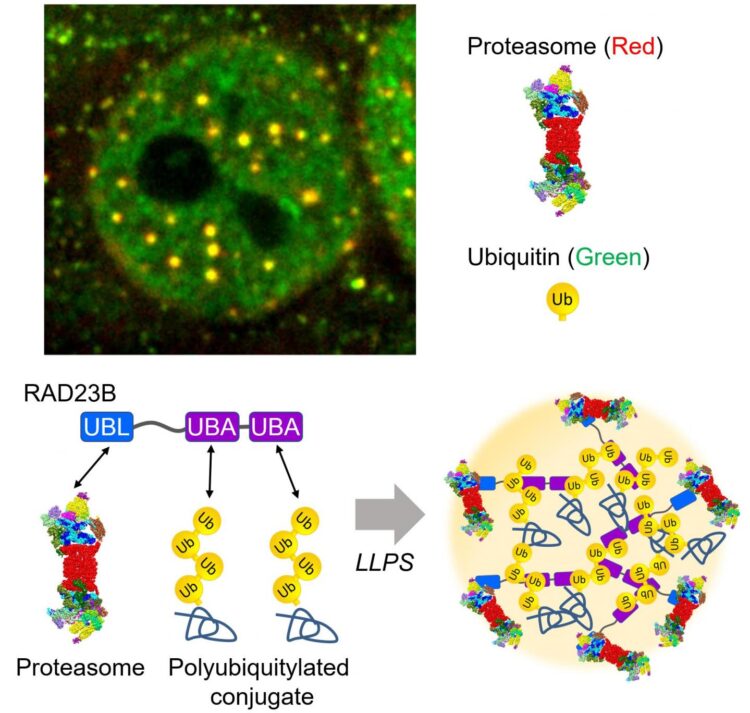
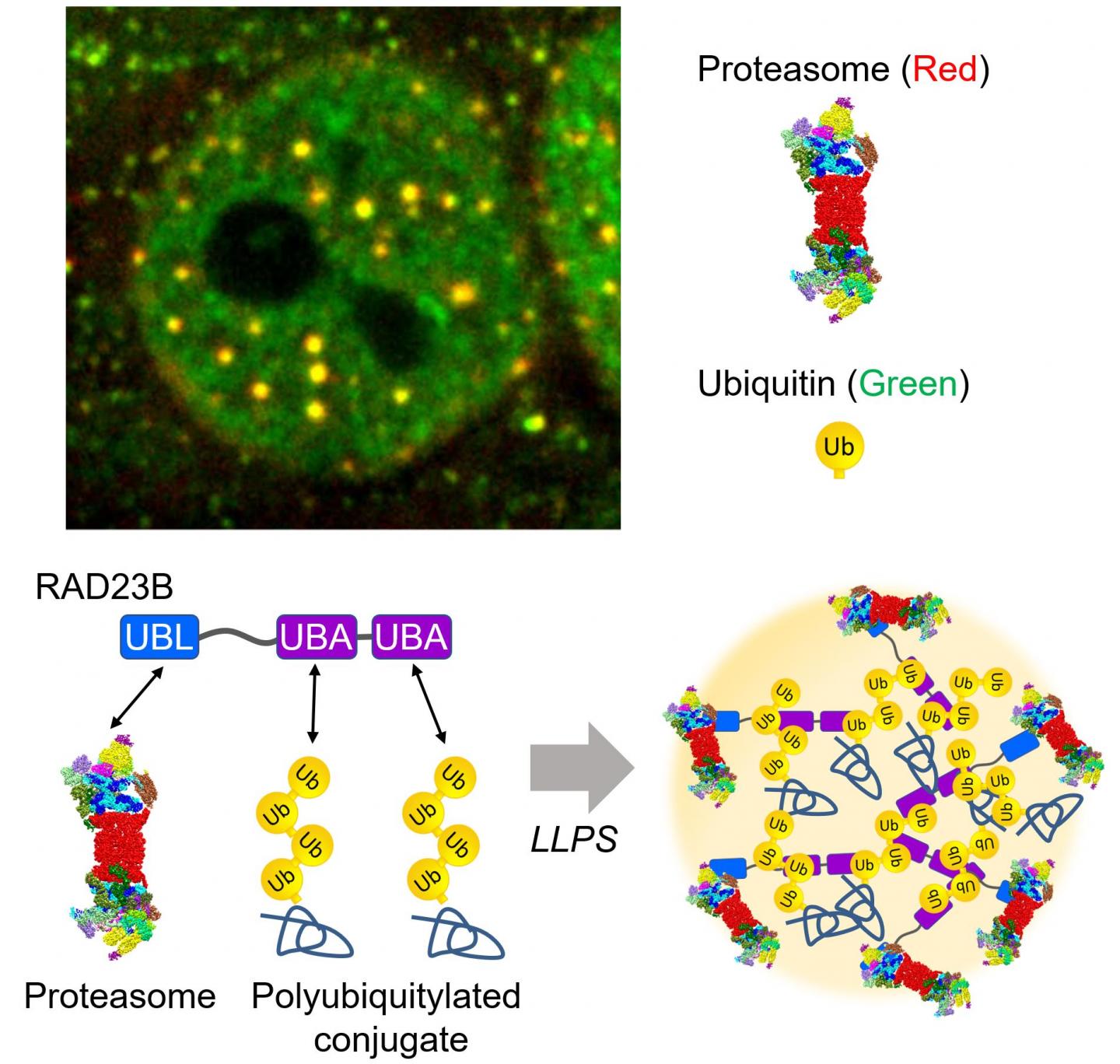
The hyperosmotic stress-induced proteasome droplets are a transient structure that disappears within a few hours after sucrose treatment. The proteasome droplets also contain ubiquitylated substrates and multiple proteasome-interacting proteins. The formation of the proteasome droplets is dependent on protein ubiquitylation and their disappearance is dependent on the activity of the proteasome, indicating that the droplets facilitate protein degradation. Acute hyperosmotic stress causes a decrease in cell volume and nucleolar stress, resulting in the failure of ribosome biosynthesis as well as accumulation of orphan ribosomal proteins as major UPS substrate in the nucleoplasm. Indeed, hyperosmotic stress induces ubiquitylation of ribosomal proteins and their degradation at the proteasome droplets. Thus, elevated levels of ubiquitylated proteins trigger the formation of the proteasome droplets.
Dr. Saeki and his colleagues further investigated how the proteasomes are recruited to this fluidic subcompartment. The research group identified RAD23B, a substrate shuttling factor of the proteasome, as a key molecule that induces LLPS of ubiquitylated clients as well as the proteasome. RAD23B has two typical ubiquitin-binding domains (UBA) and one proteasome-binding domain (UBL). The group successfully reconstituted RAD23B- and ubiquitin-containing droplets in vitro and showed that weak multivalent interactions between the RAD23B UBA domains and long polymeric ubiquitin chains drive co-phase separation. Taken together, RAD23B collects cellular ubiquitylated proteins via the UBA domains to form droplets, and then recruit the proteasomes via the UBL domain.
“This study is a good example of the interplay between ubiquitin signaling and LLPS. Given that ubiquitin mainly functions as a polymer, one of the biological meaning of the polymerization may be for LLPS. It will be greatly interesting to investigate whether other ubiquitin-binding proteins undergo phase separation.” said Dr. Saeki. “Also, it has been suggested that aggregation-prone proteins convert from liquid-like droplets to solid-like assemblies. In this context, acute hyperosmotic stress may risk irreversible accumulation of protein aggregates, especially when the proteasome activity is reduced.”
###
Media Contact
Yasushi Saeki
[email protected]
Related Journal Article
http://dx.