Credit: Purdue University image/Stephanie Venis, Hye-ran Moon and Bumsoo Han
WEST LAFAYETTE, Ind. — Pancreatic cancer has one of the worst survival rates among cancers. Patients can expect as low as a 9% chance to live for at least five years after being diagnosed.
Going back in time to observe how cells with key gene mutations interact and become invasive would help researchers better understand how the cancer starts and identify it sooner.
A pancreatic cancer “time machine” engineered by Purdue University researchers has revealed that the disease is even more unpredictable than previously thought: Cancer cells promote each other’s invasiveness when they grow together.
The study, published in the journal Small, is just the beginning of a new discovery about how pancreatic cancer evolves. Since the paper’s publication, the researchers also have found drug resistance in cancer cell types originating from two drug-sensitive ones.
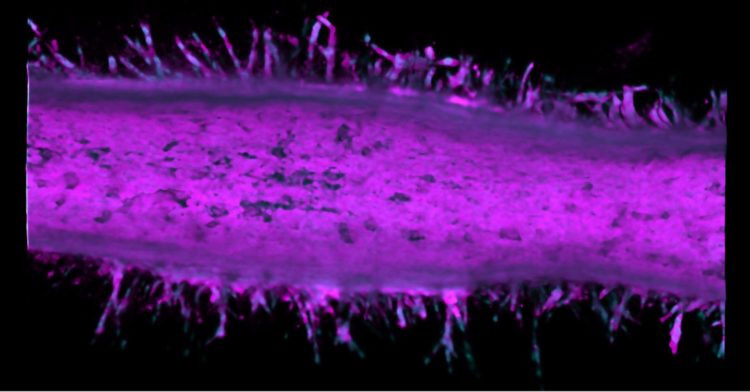
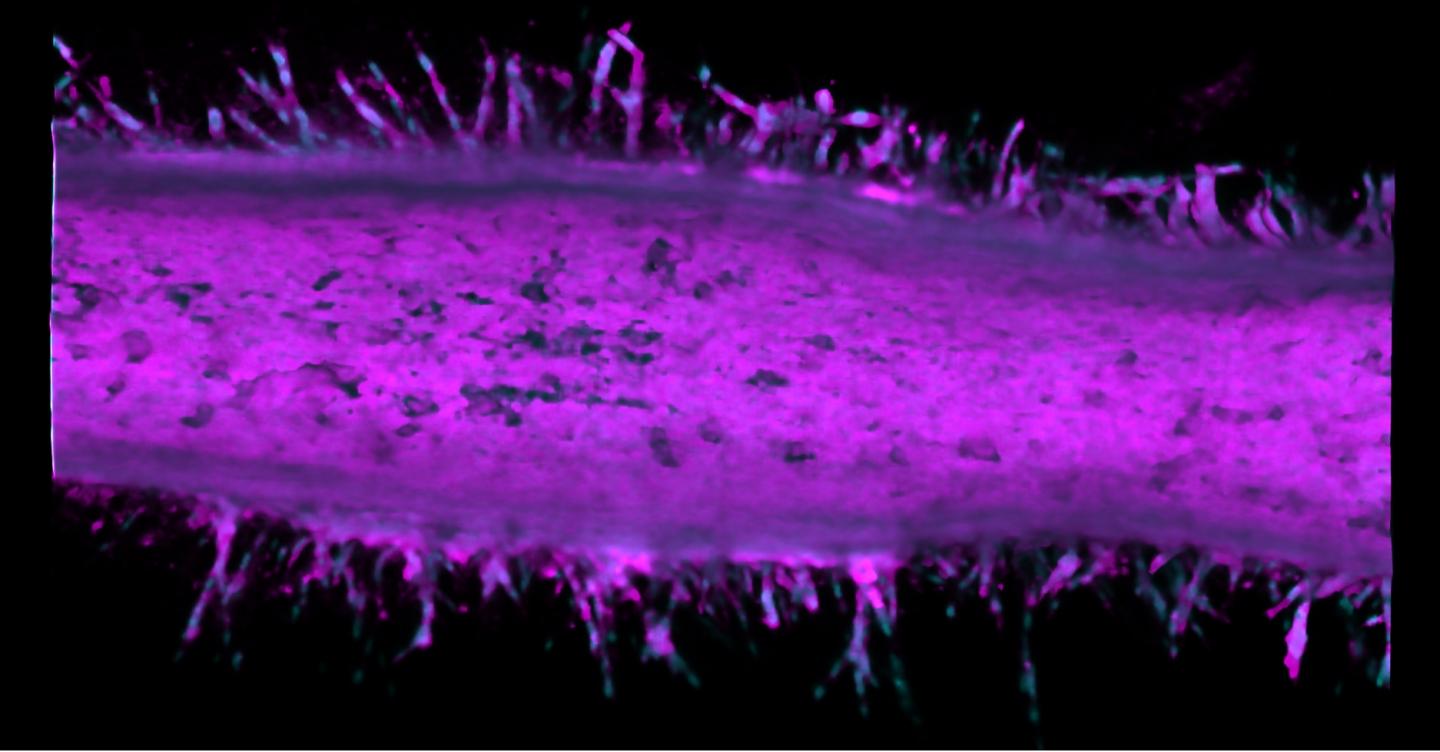
The time machine is a hollow tube of collagen that realistically mimics the microanatomy of a pancreatic duct. By injecting cancer cell lines into microfluidic channels within the artificial duct, the researchers can use the system as a model for observing how pancreatic cancer behaves over time.
Typically, it takes 10-20 years for pancreatic cancer to develop in a patient. Even in an animal model, the process is several months long. This pancreatic tumor model condenses cancer development to just two weeks.
“We can observe what happens over a long period of time. This helps us to see trends that we wouldn’t normally see,” said Bumsoo Han, a Purdue professor of mechanical engineering who builds models for studying how cancer cells move in biological systems.
The tumor model speeds up time because researchers can load in cell lines from an animal model or patient without waiting for gene mutation to happen first. The life-like structure of the tumor model allows the researchers to reconstruct the mutation as it would happen in the body.
To go back in time, the researchers just rewind footage taken by imaging equipment from the side of the artificial duct.
For this study, a group led by Stephen Konieczny, a professor of biological sciences at Purdue, developed the pancreatic cell line in a mouse model. Han’s team then loaded the cell line through the microfluidic channels of the artificial pancreatic duct. Once inside, the cell lines fill the duct and start growing.
What makes the tumor model so realistic is its shape.
“The curvature of the pancreatic duct affects the behavior of cells. We could culture these cancer cells on a petri dish, but because the dish is flat, we wouldn’t see the same behavior,” said Han, who is the program leader of the Purdue University Center for Cancer Research and has a courtesy appointment in biomedical engineering.
The researchers saw that after two different cancer cell types merged into the pancreatic tumor model device, these cells became more invasive and sprouted from the duct to form tumors.
Since cancer is technically a group of diseases, and pancreatic cancer involves four major driver mutations, Han’s team plans to further explore how each of these mutations interacts with each other. The tumor model also can be used as a prescreening tool to discover new drug targets for better drugs, Han said.
A patent has been issued for the pancreatic tumor model via the Purdue Research Foundation Office of Technology Commercialization.
###
This work was partially supported by grants from the National Institutes of Health (grants U01 HL143403, UL1 TR002529, R01 CA211098 and R01 CA124586) a Challenge Award from the Purdue University Center for Cancer Research (P30 CA023168), and the Walther Embedding Program in Physical Sciences in Oncology. Some of this research took place at the Bindley Bioscience Center, located in Purdue’s Discovery Park.
About Discovery Park
Discovery Park is a place where Purdue researchers move beyond traditional boundaries, collaborating across disciplines and with policymakers and business leaders to create solutions for a better world. Grand challenges of global health, global conflict and security, and those that lie at the nexus of sustainable energy, world food supply, water and the environment are the focus of researchers in Discovery Park. The translation of discovery to impact is integrated into the fabric of Discovery Park through entrepreneurship programs and partnerships.
ABSTRACT
A Biomimetic Tumor Model of Heterogeneous Invasion in Pancreatic Ductal Adenocarcinoma
Michael J. Bradney, Stephanie M. Venis, Yi Yang, Stephen F. Konieczny, Bumsoo Han
Purdue University, West Lafayette, IN, USA
DOI: 10.1002/smll.201905500
Pancreatic ductal adenocarcinoma (PDAC) is a complex, heterogeneous, and genetically unstable disease. Its tumor microenvironment (TME) is complicated by heterogeneous cancer cell populations and strong desmoplastic stroma. This complex and heterogeneous environment makes it challenging to discover and validate unique therapeutic targets. Reliable and relevant in vitro PDAC tumor models can significantly advance the understanding of the PDAC TME and may enable the discovery and validation of novel drug targets. In this study, an engineered tumor model is developed to mimic the PDAC TME. This biomimetic model, named ductal tumor?microenvironment?on?chip (dT?MOC), permits analysis and experimentation on the epithelial-mesenchymal transition (EMT) and local invasion with intratumoral heterogeneity. This dT?MOC is a microfluidic platform where a duct of murine genetically engineered pancreatic cancer cells is embedded within a collagen matrix. The cancer cells used carry two of the three mutations of KRAS, CDKN2A, and TP53, which are key driver mutations of human PDAC. The intratumoral heterogeneity is mimicked by co?culturing these cancer cells. Using the dT?MOC model, heterogeneous invasion characteristics, and response to transforming growth factor?beta1 are studied. A mechanism of EMT and local invasion caused by the interaction between heterogeneous cancer cell populations is proposed.
Media Contact
Kayla Wiles
[email protected]
765-494-2432
Original Source
https:/
Related Journal Article
http://dx.