Multi-omics analysis of the process establishing the mammalian body plan
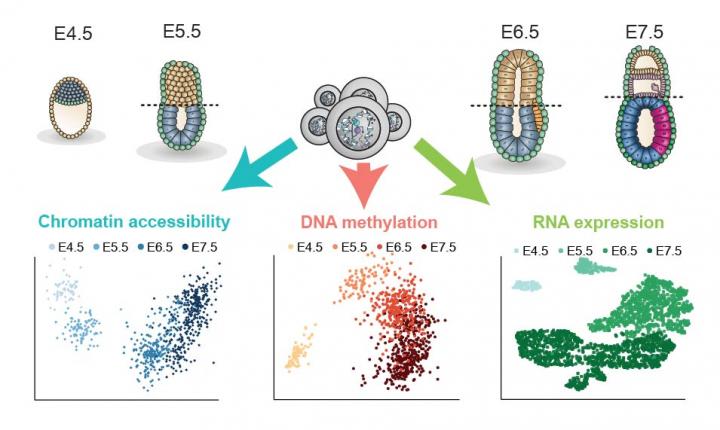
Credit: The embryo and cell images were created by Veronique Juvin from SciArtWork. Data plots were produced by Ricard Argelaguet, EMBL-EBI.
Key points:
- Uniting three different parameters in single cells taken from mouse embryos reveals more about how foundational cell identities are established in early development
- The first single-cell multi-omics analysis of gastrulation allows researchers to connect gene expression, DNA methylation and chromatin accessibility to understand the role of the epigenome in regulating cell fate decisions in early development.
- The findings propose how embryonic cells may be deviated away from a default cell state and awoken to new developmental possibilities during gastrulation, and plots the timeline of the epigenetic events controlling cell identity.
- The research brings together expertise in single-cell analysis, computational methods and machine learning from the Babraham Institute, European Bioinformatics Institute, CRUK Cambridge Institute and Wellcome – MRC Cambridge Stem Cell Institute.
Researchers at the Babraham Institute, EMBL’s European Bioinformatics Institute (EMBL-EBI), CRUK Cambridge Institute and the Wellcome – MRC Cambridge Stem Cell Institute have provided the first single-cell epigenomic analysis of gastrulation, a crucial process in early embryo development. The researchers analysed over 1,000 cells from mouse embryos to understand the epigenetic priming events preceding gastrulation and the cell fate decisions these establish. The findings, published today (Wednesday) in Nature, uncover fundamental knowledge about the processes that programme cell fate in the early embryo to generate all the organs and tissues of the body.
Just as with the construction of a building, one of the first steps before creating the upright structure is to set the foundations and the floorplan. Mammalian development is not so different. The gastrulation process takes the embryo from a ball of largely undifferentiated cells and establishes the head-to-tail and front-to-back axes and three foundational cellular layers that are each responsible for creating specific parts of the embryo.
The three layers (called germ layers) established during gastrulation are the ectoderm, mesoderm and endoderm. The ectoderm gives rise to the skin and the nervous system, the mesoderm specifies the development of several cell types such as bone, muscle and connective tissue, and cells in the endoderm layer subsequently become the linings of the digestive and respiratory system, and form organs such as the liver and the pancreas.
Building on exciting progress in the use of cutting-edge single-cell and computational techniques to understand early development, the researchers used two pioneering methods to collect and analyse the gastrulation data. The first was a technique developed at the Babraham Institute called scNMT-seq (single-cell Nucleosome, Methylome and Transcriptome sequencing). This was used to obtain multiple biological read-outs from 1,105 single cells taken from mouse embryos at different development stages spanning the gastrulation process. In each cell, the researchers assessed gene expression activity, DNA methylation and chromatin accessibility to chart how these changed as cells went through the gastrulation process.
The second was a computational method developed by researchers at EMBL-EBI called Multi-Omics factor Analysis (MOFA). This machine-learning approach, originally developed for personalised medicine, allowed the researchers to unite the three streams of biological information profiled from each single cell.
Ricard Argelaguet, a PhD student at the EMBL-EBI co-supervised by John Marioni and Oliver Stegle, said “MOFA offers a principled approach for combining high-dimensional multi-omics data. It helped us to identify the elements in the genome that are associated with cell fate commitment as well as enabling us to understand how different molecular features interact with one another throughout gastrulation.”
The researchers found that the three germ layers (ectoderm, mesoderm and endoderm) showed differences in the timing of epigenetic events in a hierarchical way. The analysis demonstrated that ectoderm cells were primed epigenetically at an earlier developmental stage. This finding may explain the existence of a pre-programmed default developmental path specifying skin and brain identities. Epigenetic events occurring later in mesoderm and endoderm cells may act to actively divert these cells away from this default path by making the cells receptive to signals promoting other cell identities.
“Through analysing the timeline of events, we identified that the diversification of the three gastrulation layers was mainly driven by epigenetic events affecting germ layer specific enhancers.” said Dr Stephen Clark, lead researcher and one of the paper’s four joint first authors. “We found that the epigenome of the ectoderm layer was established much earlier in development than the other two, even though all three cell types arise at a similar time.”
Professor Wolf Reik, Head of the Epigenetics Programme at the Babraham Institute, said “This is the first comprehensive application of the single-cell method developed here at the Institute to a biological question and gives a new view on how cell fate is established. Our findings develop our understanding of the role of the epigenome in defining cell fate commitments at different stages of development, with important implications for stem cell biology and medicine. It is very enjoyable to see how the multidisciplinary research community that has come together in this project is now sharing the success of their efforts.”
Dr John Marioni, Group Leader at the EMBL-EBI and the CRUK Cambridge Institute, said “The ability of a cell to commit to a specific fate requires integration of a complex array of different molecular signals. Analogously, understanding this process requires combining cutting-edge experimental and computational tools, as exemplified in this work. Importantly, our data and analyses provide a blueprint for how the epigenome might regulate cell fate choice in other contexts, providing an exciting launch pad for future studies.”
A newly announced research initiative, the Wellcome-funded Human Developmental Biology Initiative, will utilise the same single-cell methods to analyse cells from human embryos. The initiative, which involves several of the paper’s authors (Professor Wolf Reik, Dr Gavin Kelsey, Dr Peter Rugg-Gunn, each a group leader at the Babraham Institute, and Professor Bertie Göttgens, Principal Investigator, Wellcome – MRC Cambridge Stem Cell Institute) might provide insight as to whether the same epigenetic mechanisms of control work similarly in humans.
Single-cell multi-omics analysis is also likely to deliver significant impacts for human healthcare in future years. A pan-European research initiative, LifeTime, is bringing together life science experts with leaders in the pharma, clinical medicine and technology industries to map how innovation and cutting-edge technologies (including single-cell multi-omics methods) can be united to revolutionise healthcare.
###
Media Contact
Louisa Wood
[email protected]
44-012-234-96230
Related Journal Article
http://dx.




