Credit: University of Cologne (Germany) & USWR/Tehran (Iran)
An international research team including scientists from the University of Cologne, Germany, numerous Iranian universities and the University of Sydney, Australia, has shown that today’s Iranian population is composed of partially highly heterogeneous ethnic groups, exhibiting a high degree of genetic variation. In many cases, their source goes back many thousands of years. The results, obtained from the first genome-wide genetic characterization of the Iranian population by this team, appeared in PLOS Genetics under the title ‘Distinct genetic variation and heterogeneity of the Iranian population’.
In this project, initiated by Professor Michael Nothnagel from the Cologne Center for Genomics (CCG) at the University of Cologne and Professor Hossein Najmabadi from the Genetics Research Center (GRC) at the University of Social Welfare and Rehabilitation Science in Tehran, their groups cooperated with numerous other universities in Iran, while Professor Barbara Helwing from Sydney provided expertise on Iran’s historical background.
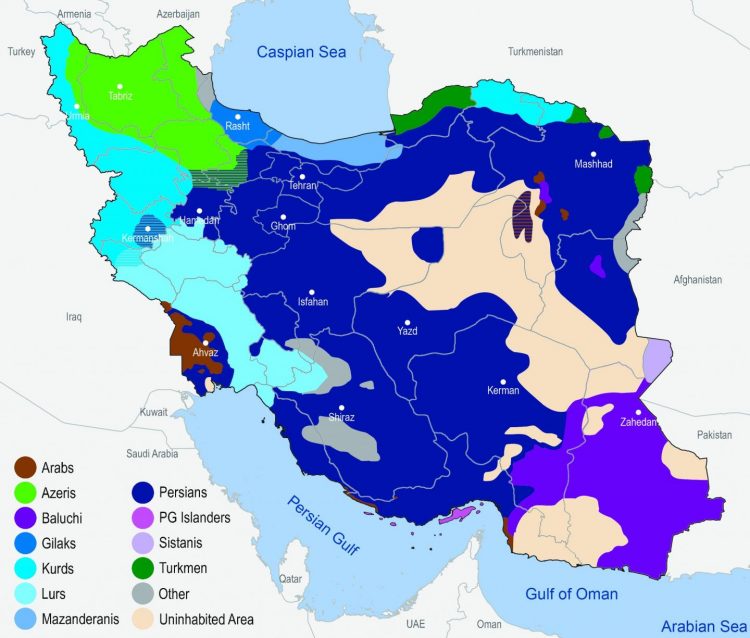
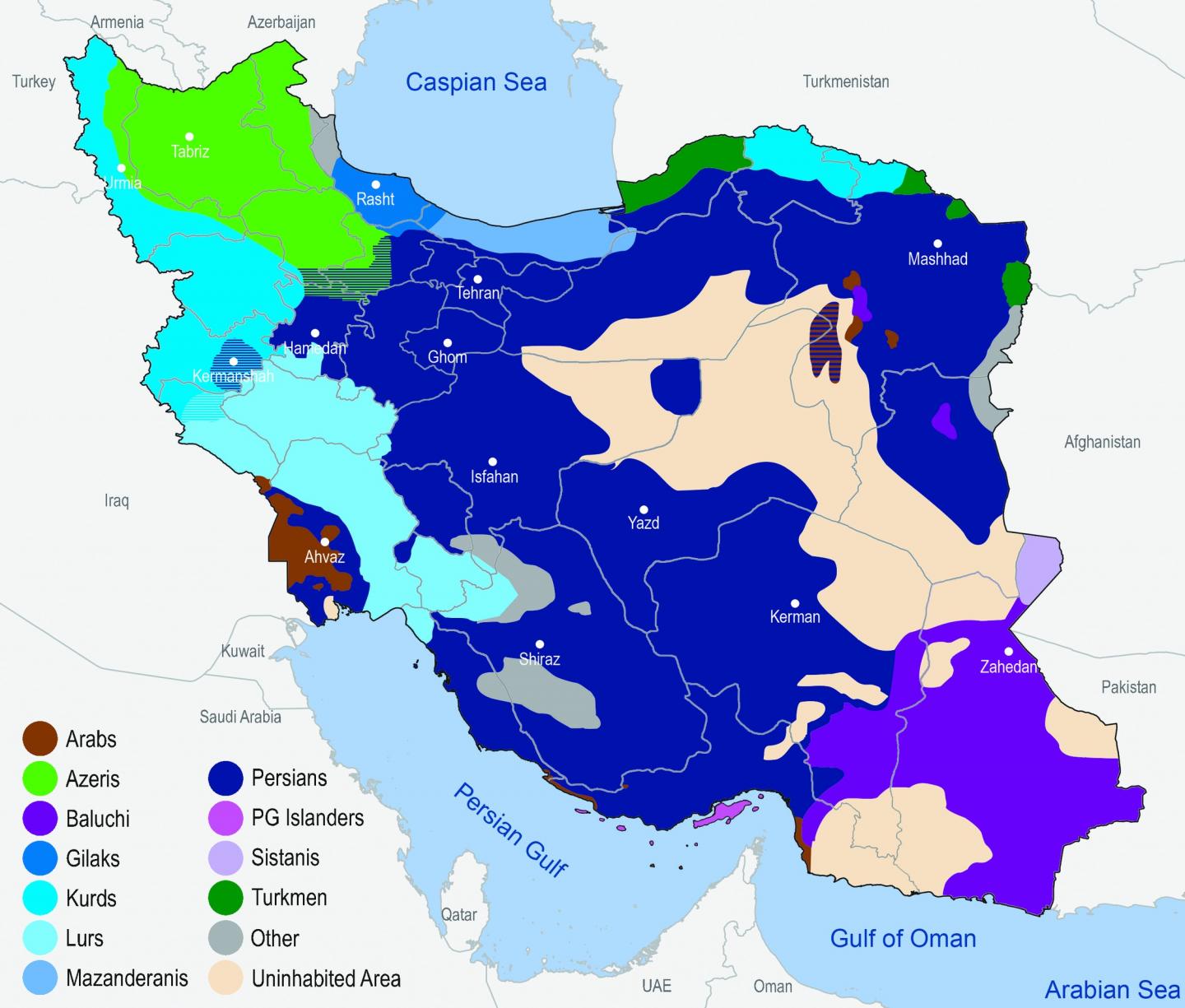
The researchers analysed the genetic data of 1,021 volunteers whose parents and grandparents identified themselves as belonging to one of eleven selected Iranian ethnic groups, including large groups such as Iranian Persians and Azeri, but also smaller ones like Arabs, Baluchi, Gilaki and Kurds. These volunteers were sampled all over Iran. They found out that Iranian Persians and Kurds, for example, exhibit high in-group genetic variation which is larger than that of, for example, Germans. However, the entire gene pool has remained largely unchanged over at least the past 5,000 years, but probably rather the past 10,000 years. To put this in perspective: Today’s German population has likely retained only about 10 to 20 percent of the genetic constitution of the hunters and gatherers who populated western and central Europe 10,000 years ago. Furthermore, Britons and North Italians are genetically more similar than some ethnic groups in Iran. ‘This was somewhat surprising,’ Michael Nothnagel said. ‘Until recently, many scientists had assumed genetic variation across present-day Iranians to be rather homogeneous.’
In addition, the authors put the genomic data they collected in relation to published data from other living human populations and from almost 800 excavated specimens originating from individuals who lived in the region and beyond thousands of years ago. Michael Nothnagel remarked: ‘The genetic information from Iran is particularly valuable because it fills a void of population-representative, genome-wide data for a large population in an important region of the world.’ The generated data sets and the patterns they reveal will be valuable for follow-up research on rare and common genetic diseases and help to elucidate past migration movements.
Iran has been at the crossroads of migrations since modern humans’ dispersal out of Africa and played an important cultural and political role in Western and Central Asia in the past millennia. However, despite its size, geographic location and past cultural influence, the country has largely been a blind spot for human population genetic studies, resulting in sparse genetic information on the population.
In the past millennia, Iran has repeatedly received migratory influx: Indo-European language speakers settled there, Arabs entered the lands in the 7th century, and later Turkic-speaking people from Central Asia joined the population. As a result, today’s Iranian population comprises numerous ethnic, religious and linguistic groups that admixed to various degrees.
For example, while Iranian Kurds show strong genetic overlaps with Persians, Lurs and other groups, indicating a common background and little admixture at most, Persian Gulf Islanders appear to have repeatedly received an influx of groups from outside Iran. ‘This is consistent with historical reports of ongoing sea trade over the past millennia,’ said Nothnagel. ‘Also, some groups adopted the languages of newly arrived groups. This shows that the latter did not simply displace local populations, but rather mixed with them.’ Combining archaeological finings with the latest technologies in genome characterization, the study provided systematic evidence of these migration and settlement patterns.
###
Media Contact
Michael Nothnagel
[email protected]
Related Journal Article
http://dx.