Synthetic biologists add tunable, analog-to-digital converter to their toolkit
Credit: Rice University
HOUSTON — (April 18, 2019) — Synthetic biologists have added high-precision analog-to-digital signal processing to the genetic circuitry of living cells. The research, described online today in the journal Science, dramatically expands the chemical, physical and environmental cues engineers can use to prompt programmed responses from engineered organisms.
Using a biochemical process called cooperative assembly, Caleb Bashor of Rice University, Ahmad “Mo” Khalil of Boston University (BU) and colleagues from MIT, Harvard, the Broad Institute and Brandeis University engineered genetic circuits that were able to both decode frequency-dependent signals and conduct dynamic signal filtering.
“You can think about cooperativity as the same type of signal-processing feature that gives you an analog-to-digital converter, a device that takes something that’s basically linear and turns it into something switchlike,” said Bashor, co-lead author of the study and an assistant professor of bioengineering in Rice’s Brown School of Engineering.
Synthetically engineering cooperative assembly allowed the researchers to perform the type of combinatorial signal processing that cells naturally and elegantly do to accomplish intricate tasks, like those in embryonic development and differentiation.
“This work is a tour de force of synthetic biology that addresses a major question in how cells process information at the DNA level,” said Tom Ellis, Reader in Synthetic Genome Engineering in the department of bioengineering at Imperial College London, who was not involved in the study. “It’s well known that nature has perfected very powerful information processing with only a small number of parts, but deconvoluting precisely how this works is virtually impossible in human cells due to their complexity. By recreating the way human cells process information at the DNA level, but in a simple yeast cell model with synthetic parts, they have been able to recreate complex signaling from first principles. This is an excellent example of how thinking like an engineer can unlock a new way to answer major biology questions.”
In nature, cells often have to make black-and-white decisions based on information that’s gray. For example, imagine a cell has a gene that allows it survive in a highly acidic environment, but it takes a good deal of energy to activate that gene and get the protection. Through billions of years of natural selection, cells that activate the gene too early or too late get outcompeted by those that make the decision at the optimum time to both ensure survival and expend the least amount of energy.
“That type of precision is a desirable property to have in synthetic circuits, too,” said Bashor, who joined Rice in 2018 and began the project several years earlier during a postdoctoral stint at BU. “Nature often does it through a process called cooperative self-assembly, where several proteins called transcription factors self-assemble into a larger complex. Only when they come together is the switch thrown.”
Bashor, Khalil and colleagues engineered cooperative self-assembly by inventing a modular system of synthetic protein components that can assemble into complexes of varying size. In this system, engineered cells are programmed to produce assembly components in response to whatever input the engineers wish to use to activate the circuit. For example, in their experiments, Bashor, Khalil and colleagues programmed yeast to respond to two different drugs that were administered in varying concentrations via a microfluidic device.
In this way, the concentration of component molecules produced inside the yeast rose and fell in response to the analog input — the concentration of drugs in the test chamber.
“Basically, these components bind to one another with extremely weak interactions,” Bashor said. “But all of those weak interactions add up, in a bigger complex, to something that’s really tight. So, when there’s very few of them floating around, they won’t form the complex. And when they reach a critical concentration, they see each other, and they can basically come together and form the complex.”
The sharpness of a response — one that happens quickly at precisely the intended time — is key for digital precision. Bashor and Khalil designed activation complexes that contained as few as two transcription-factor components and as many as six, and their experiments showed that the larger the complex, the sharper the critical response.
“Engineering this type of response into transcription factors was central for allowing us to program cells to perform a diverse array of complex functions, such as Boolean logic, time-dependent filtering and even frequency decoding,” said Khalil, the corresponding author on the study.
Bashor said the bulk of the four-year project was spent refining a predictive model that can guide other engineers in using the system to design analog-to-digital converters that can respond as intended even to multiple incoming signals.
To demonstrate this aspect of the work, the team designed and demonstrated signal-processing circuits reminiscent of microelectronics, including low-pass filters that responded only to low-frequency drug inputs and band-stop filters that were activated only at high frequencies.
“Our work shows how the nonlinearity of transcription factor complexes can be used to engineer signal processing in synthetic gene circuits, expanding their functionality and real-world utility,” said synthetic biologist and study co-author James Collins, who holds joint appointments at MIT, Harvard and the Broad Institute.
Going forward, Bashor’s Rice lab plans to use the analog-to-digital converter and other synthetic gene circuits to explore and manipulate the regulatory programs that guide immune and stem cell functions with an eye on developing transformational cell-based therapeutics from engineered human cells.
###
Additional co-authors include study co-lead author Nikit Patel, formerly of BU and now with Harvard Medical School; Ali Beyzavi, formerly of BU and now with MIT; Sandeep Choubey, formerly of Brandeis and now with the Max Planck Institute for the Physics of Complex Systems in Dresden, Germany; and Jané Kondev of Brandeis.
The research was supported by the National Science Foundation, the Defense Advanced Research Projects Agency and the National Institutes of Health.
High-resolution IMAGES are available for download at:
https:/
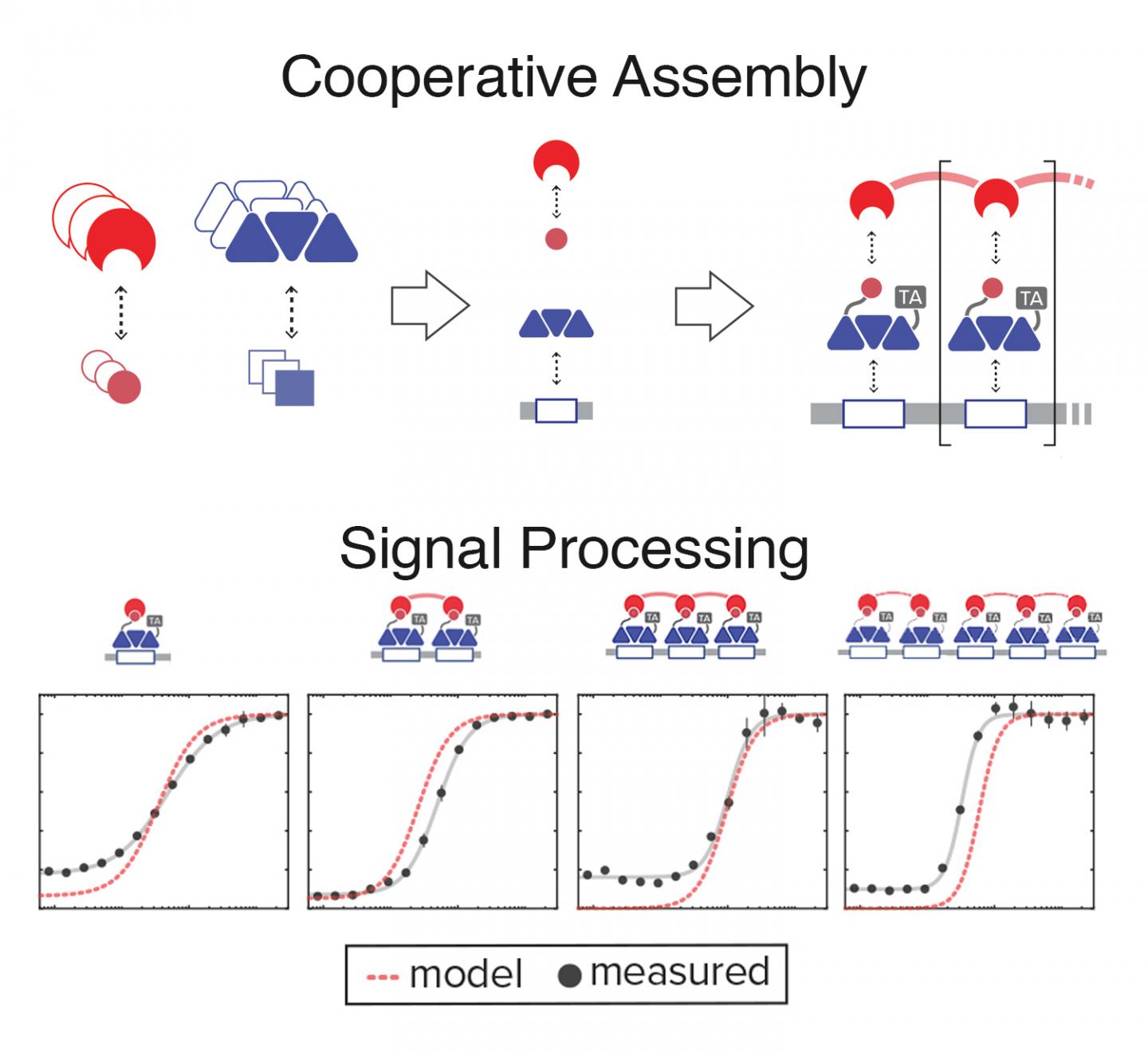
CAPTION: Synthetic biologists designed a system for engineering cells that respond digitally to analog signals. Cells are pre-programmed to produce transcriptional protein components (top left) in proportion to the strength of an incoming signal. Upon reaching a critical concentration, components self-assemble (top right) into a protein complex that initiates transcription of a pre-programmed target gene. The more modules a self-assembled transcription complex contains (bottom, left to right), the more sharply it responds to the critical threshold. (Image courtesy of Rice University)
http://news.
CAPTION: Caleb Bashor is an assistant professor of bioengineering at Rice University (Photo by Jeff Fitlow/Rice University)
http://www.
CAPTION: Ahmad “Mo” Khalil is an assistant professor of biomedical engineering at Boston University (Photo by Mike Pecci for Boston University)
The DOI of the Science First Release paper is: 10.1126/science.aau8287
A copy of the paper is available at: https:/
George R. Brown School of Engineering: engineering.rice.edu
Rice’s Department of Bioengineering: bioengineering.rice.edu
Related research from Rice:
DIY brings high throughput to continuous cell culturing — June 11, 2018
Switch-in-a-cell electrifies life — Dec. 17, 2018
Rice U. announces $82 million in strategic research initiatives — Oct. 16, 2018
Models give synthetic biologists a head start — Aug. 14, 2018
?Synthetic biologists engineer inflammation-sensing gut bacteria — April 5, 2017
New tools advance bio-logic — Aug. 4, 2014
This release can be found online at news.rice.edu.
Follow Rice News and Media Relations via Twitter @RiceUNews.
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,962 undergraduates and 3,027 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 2 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance.
Media Contact
Jade Boyd
[email protected]
Related Journal Article
http://dx.




