Study upends ideas inferred from nonwoody plants
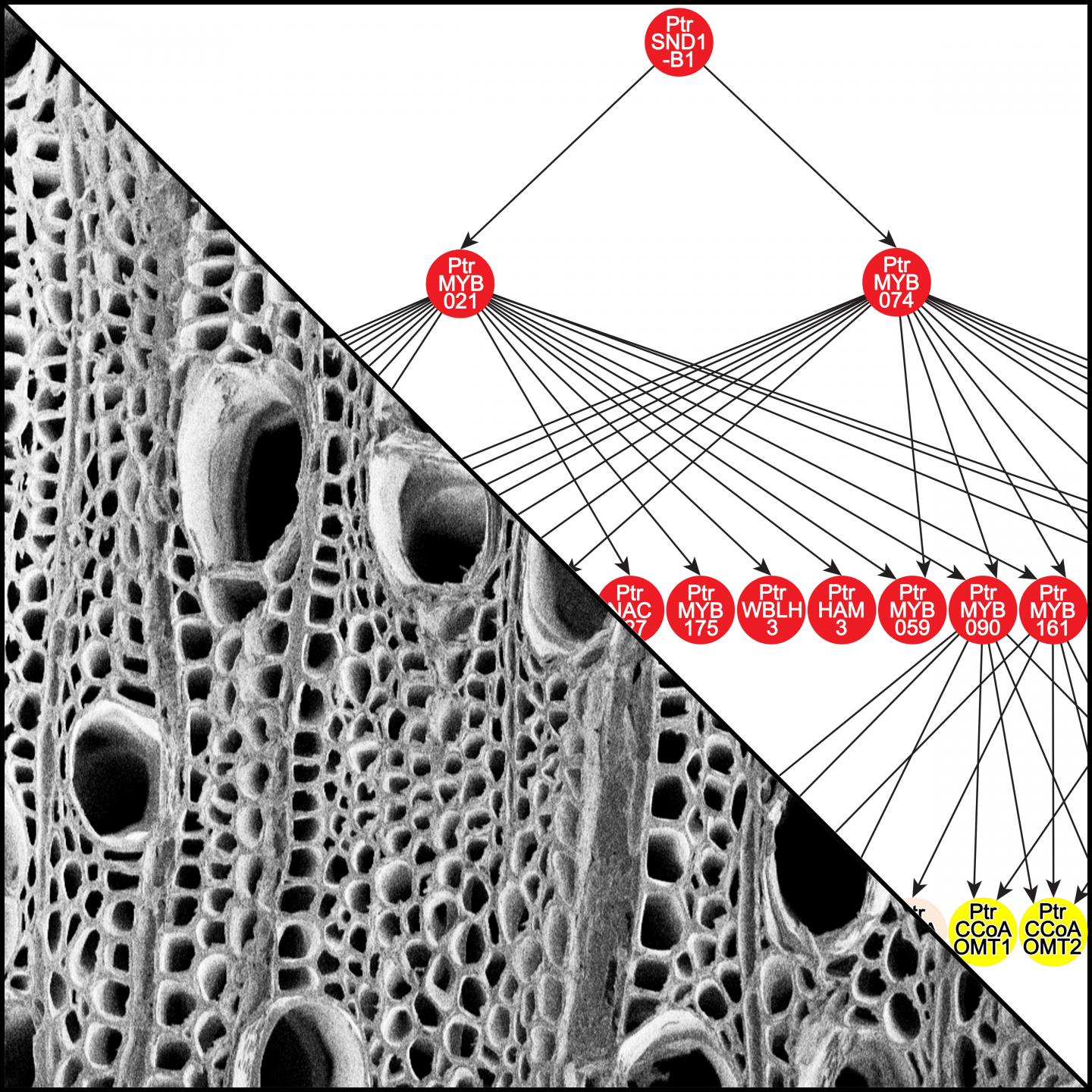
Credit: Illustration by Jack P. Wang and Ilona Peszlen.
North Carolina State University researchers have uncovered how a complex network of transcription factors switch wood formation genes on and off. Understanding this transcriptional regulatory network has applications for modifying wood properties for timber, paper and biofuels, as well as making forest trees more disease- and pest-resistant.
“We’re building a complete story, so to speak, of how wood formation functions – all the intricate components, how they interact and how they fit together to regulate wood formation inside the cell walls of woody plants,” says Jack Wang, assistant professor in the College of Natural Resources and co-lead author of a Plant Cell article about the work.
Researchers with NC State’s Forest Biotechnology Group used transgenic black cottonwood (Populus trichocarpa), a species they’ve studied intensively, to identify interactions in a transcriptional regulatory network directed by a key transcription factor, PtrSND1-B1. The researchers documented four levels of interactions in the network, from DNA to enzyme levels. The work is an extension of two previous studies, in which functions of PtrSND1-B1 were discovered using a wood-forming cell system. These early studies were published in PNAS and Plant Cell by co-authors Quanzi Li, Ying-Chung Lin and Wei Li of the Forest Biotechnology Group, led by Vincent Chiang.
“Transcription factors – a complex network of them – regulate which wood formation genes are turned on or off,” Wang says. “Essentially these are high-level regulatory switches.
“Understanding this network allows us to identify single switches inside that complex network of transcription factors that could simultaneously control multiple wood-forming genes. Instead of working with one, two or three genes at a time, which is our current limit, plant biologists could work with tens of genes at a time.”
The new study upends ideas about transcriptional regulatory networks inferred from work with nonwoody species, such as Arabidopsis thaliana, a model plant.
“This network of transcription factor regulation in woody tissues is almost completely different from the regulatory processes in Arabidopsis and other plants,” says Hao Chen, co-lead author of the article and a postdoctoral researcher at NC State.
“Of 57 regulatory interactions we identified, 55 were specific to woody plant tissue, showing that herbaceous plants like Arabidopsis cannot stand in for woody plants.”
The study provides an extensive look at a transcriptional regulatory network in woody plants. Researchers’ goal is to provide a toolkit for building trees with specific properties needed for commercial timber, paper, biofuel production and conservation needs.
Plant biologists tested 42 of the interactions they found in lines of transgenic black cottonwood, verifying the function of about 90 percent. The network revealed which genes are common targets for specific transcription factors. As a result, researchers found nine new protein-protein interactions involved in forming lignin, a component in the cell wall that gives wood its strength and density.
Wang says several recent studies show that lignin is related to disease and insect resistance in trees, a major concern. A 2012 U.S. Forest Service report estimated that 7 percent of the nation’s forests are in jeopardy of losing more than a quarter of their tree vegetation by 2027. The amount of threatened vegetation rose by 40 percent in just six years.
“Studies like this that look at lignification and wood formation will have great value in helping to understand how trees can be made to be more robust and to improve forest health in general,” Wang says.
###
This work was supported in part by the U.S. Office of Science (Biological and Environmental Research), Department of Energy Grant DESC000691.
Note: An abstract of the paper follows.
“Hierarchical Transcription-Factor and Chromatin Binding Network for Wood Formation in Populus trichocarpa”
Authors: Hao Chen, Jack P. Wang, Huizi Liu, Huiyu Li, Ying-Chung Jimmy Lin, Rui Shi, Chenmin Yang, Jinghui Gao, Chenguang Zhou, Quanzi Li, Ronald R. Sederoff, Wei Li, Vincent L. Chiang
Published: Plant Cell
DOI: https:/
Abstract: Wood remains the world’s most abundant and renewable resource for timber and pulp, and an alternative to fossil fuels. Understanding the molecular regulation of wood formation can advance the engineering of wood for more efficient material and energy productions. We integrated a Populus trichocarpa wood forming cell-system with quantitative transcriptomics and chromatin-binding assays to construct a transcriptional regulatory network (TRN) directed by a key transcription factor (TF), PtrSND1-B1. The network consists of 4 layers of TF-target gene interactions with quantitative regulatory effects, describing the specificity of how the regulation is transduced through these interactions to activate cell-wall genes (effector genes) for wood formation. PtrSND1-B1 directs 57 TF-DNA interactions through 17 TFs transregulating 27 effector genes. Of the 57 interactions, 55 are novel. We tested 42 of these 57 interactions in 30 genotypes of transgenic P. trichocarpa and verified that ~ 90% of the tested interactions function in vivo. The TRN reveals common transregulatory targets for distinct TFs, leading to the discovery of 9 novel TF protein-complexes (dimers and trimers) implicated in regulating the biosynthesis of specific types of lignin. Our work suggests that wood formation may involve regulatory homeostasis determined by combinations of TF-DNA and TF-TF (protein-protein) regulations.
Media Contact
D’Lyn Ford
[email protected]
Original Source
https:/
Related Journal Article
http://dx.