Scientists from the Federal University of Rio de Janeiro explored mRNA and protein databases to unravel hidden meanings of the genetic code
Credit: Rodolfo Carneiro
Today, thousands of databases with biological data are publicly available. They include data on gene and protein sequences and detailed measurements of different cellular parameters, such as the exact quantities of all proteins produced and degraded by a given cell in various experimental conditions. Brazilian researchers explored mRNA and protein public databases and found out how gene sequence choice can predict different aspects of protein synthesis, such as protein production efficiency. The study, published in Nucleic Acids Research, could help the development of new biotechnological applications of genes and proteins.
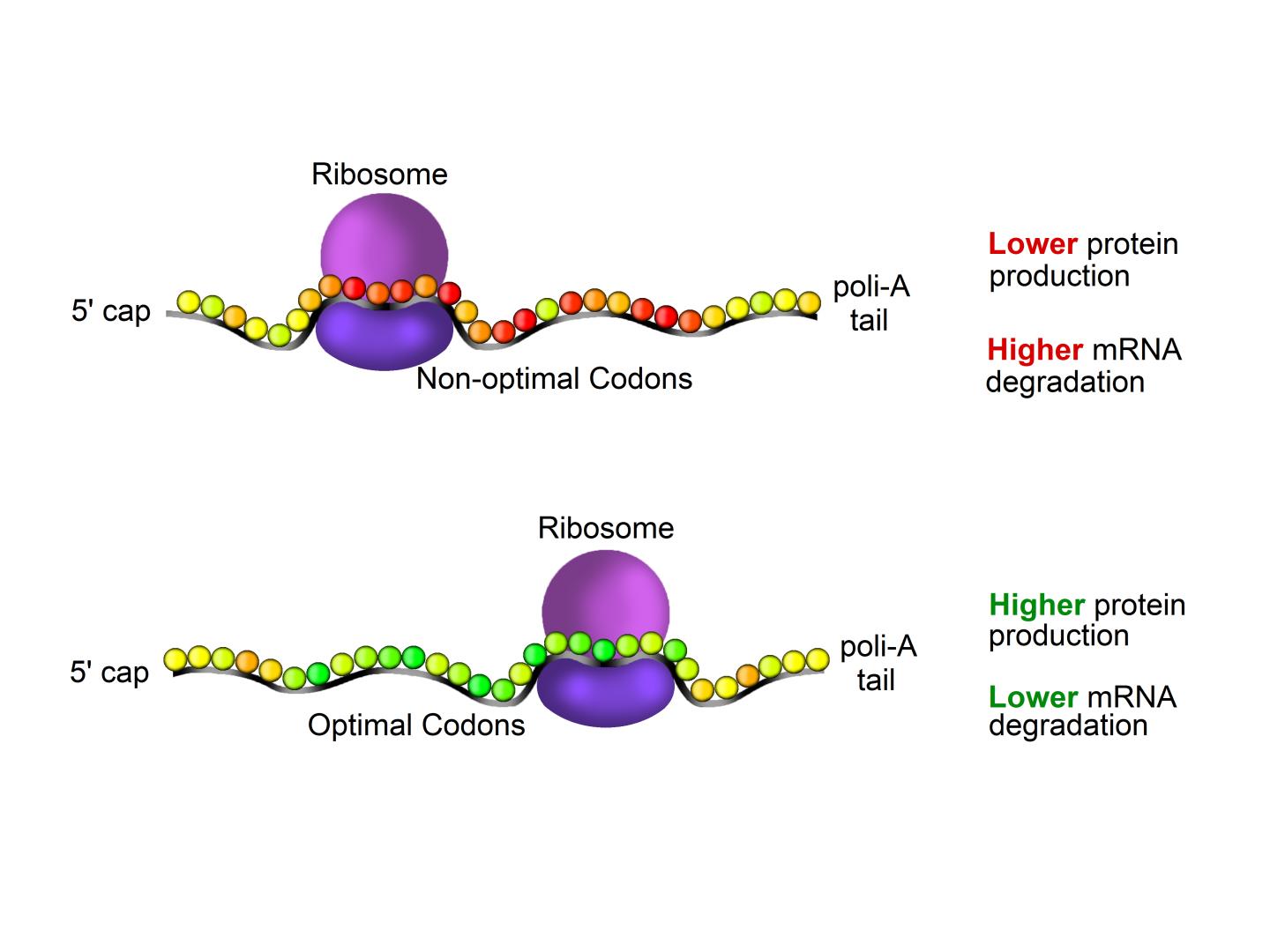
The genetic information contained in the cell nucleus in the form of DNA is copied in messenger RNAs (mRNAs). Different from the DNA, mRNAs are dynamic and unstable molecules that leave the nucleus and are translated by the ribosomes, the molecular machines able to convert a sequence of nucleotides that make RNA (and DNA) into a sequence of amino acids that form proteins. Each amino acid corresponds to one or more combinations of 3 nucleotides – or codon. Because the same amino acid can be translated from different codons, the genetic code is described as degenerate (or redundant).
Scientists already know that even though the same protein can be produced from alternative gene sequences, some combinations result in higher protein yields. They also know that optimal codons and non-optimal codons can decrease or enhance mRNA degradation, respectively. Different groups have measured mRNA production and degradation rates, but, surprisingly, there are many deviations in the data.
Brazilian scientists synthesized apparently disparate pieces of data and extended our knowledge of how gene sequence choice can predict different aspects of protein synthesis, such as mRNA stability and production efficiency. A research group led by Fernando Palhano and Tatiana Domitrovic at the Federal University of Rio de Janeiro used a metric derived from mRNA codon composition to compare the existing data to different cellular parameters. They found that this metric correlated well with protein abundance and protein production efficiency, indicating the most coherent mRNA decay datasets. Their work reiterated that mRNA degradation is somehow connected to protein production efficiency. “Even proteins needed in high levels under specific conditions, such as stress response, have their gene sequence optimized for efficient translation”, says Fernando Palhano.
Fernando and Tatiana worked with Rodolfo Carneiro and other colleagues, who identified a group of low abundance proteins coded by a non-optimal subset of codons. As they show in their paper published in Nucleic Acids Research, codon choice is vital not only to guarantee high protein production but also to tune down the output of proteins that should be produced in minimum amounts, such as regulatory proteins.
The amount of protein produced in a cell is crucial to maintaining the organism function – “Many human diseases are caused by inefficient or unbalanced protein production, such as cystic fibrosis and cancer”, says Tatiana. She adds that “from a practical perspective, understanding the relationship between the genetic sequence and protein production can have a profound effect both on medicine and bioengineering”.
The authors note that many “silent” DNA mutations, that is, mutations that alter the codon sequence, but not the coded amino acid, can lead to significant modifications on protein production rates, which could lead to disease. By carefully selecting the gene sequence one can finely tune the protein production and boost biotechnological applications of genes and proteins.
###
The paper entitled “Codon stabilization coefficient as a metric to gain insights into mRNA stability and codon bias and their relationships with translation” is published online in Nucleic Acids Research.
Media Contact
Fernando Palhano
[email protected]
55-219-991-89941
Related Journal Article
http://dx.




