Credit: Lees et al. 2019
Jan 24, 2019 — Differences in genetic diversity among bacterial pathogens correlate with clinically important factors, such as virulence and antimicrobial resistance, prompting the need to identify clusters of similar bacterial strains. However, current bacterial clustering and typing approaches are not suitable for real-time pathogen surveillance and outbreak detection.
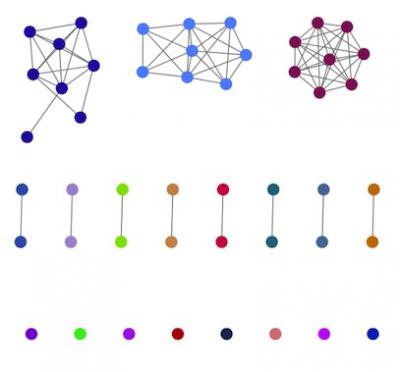
In a study published today in Genome Research, researchers developed PopPUNK (Population Partitioning Using Nucleotide K-mers), a computational tool for analyzing tens of thousands of bacterial genomes in a single run, up to 200-fold faster than previous methods. Using k-mers, short sections of DNA length k, this software enables rapid estimation of the proportion of k-mers present in one genome that are also shared by another. Differences in k-mer content between genomes may represent changes to individual bases in otherwise similar stretches of DNA or differences in gene content. By calculating these relationships across isolates, the population structure of a species can be efficiently estimated.
Importantly, PopPUNK applies a machine learning method that enables easy identification of emerging strains in a population. Using a previously published data set of E. coli isolates collected over a ten-year study, PopPUNK was able to efficiently classify the prevalence of different strains in the population each year and identify the emergence of antibiotic-resistance strains over time.
Researchers envision PopPUNK will expedite the identification of bacterial strains as the scale of bacterial genomes being sequenced increases and, importantly, allow public health agencies to quickly identify outbreak strains that pose a public health risk.
###
Researchers from New York University School of Medicine, Wellcome Sanger Institute, University of Helsinki, University of Cambridge, and Imperial College London contributed to this work. The study was funded by grants from the United States Public Health Service, Wellcome, Bill and Melinda Gates Foundation, European Research Council, and the Royal Society.
The authors are available for more information by contacting Greg Williams, NYU School of Medicine media contact; +1-212-404-3533) or Ryan O’Hare, Imperial College London communications and public affairs office; +44-(0)20-7594-2410). Interested reporters may obtain copies of the manuscript via email from Dana Macciola, Administrative Assistant, Genome Research; +1-516-422-4012).
About the article:
The manuscript will be published online ahead of print on 24 Jan 2019. Its full citation is as follows: Lees J, Harris S. Tonkin-Hill G, Gladstone R, Lo S, Weiser J, Corander J, Bentley S, Croucher N. 2019. Fast and flexible bacterial genomic epidemiology with PopPUNK. Genome Research doi: 10.1101/gr.241455.119
About Genome Research:
Launched in 1995, Genome Research is an international, continuously published, peer-reviewed journal that focuses on research that provides novel insights into the genome biology of all organisms, including advances in genomic medicine. Among the topics considered by the journal are genome structure and function, comparative genomics, molecular evolution, genome-scale quantitative and population genetics, proteomics, epigenomics, and systems biology. The journal also features exciting gene discoveries and reports of cutting-edge computational biology and high-throughput methodologies.
About Cold Spring Harbor Laboratory Press:
Cold Spring Harbor Laboratory Press is an internationally renowned publisher of books, journals, and electronic media, located on Long Island, New York. Since 1933, it has furthered the advance and spread of scientific knowledge in all areas of genetics and molecular biology, including cancer biology, plant science, bioinformatics, and neurobiology. The Press is a division of Cold Spring Harbor Laboratory, an innovator in life science research and the education of scientists, students, and the public. For more information, visit our website at http://cshlpress.
Media Contact
Dana Macciola
[email protected]
Related Journal Article
http://dx.