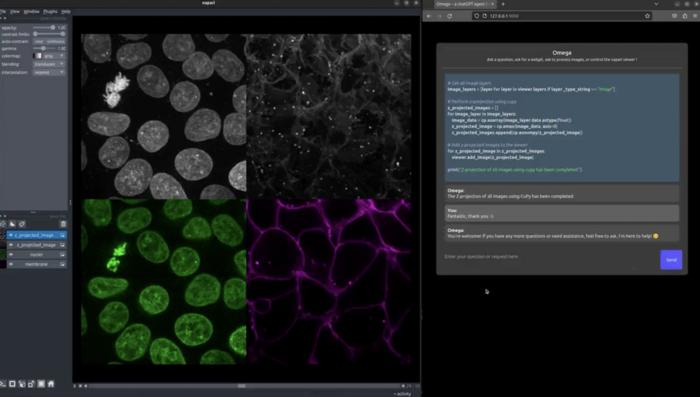
In a new research article, scientists at Chan Zuckerberg Biohub San Francisco (CZ Biohub SF) describe Omega, an open-source software tool that significantly advances the field of bioimage analysis. Omega harnesses the power of large language models (LLMs) to enable scientists to process and analyze biological images through natural language conversations rather than having to issue formal commands or write code.
Credit: Chan Zuckerberg Biohub San Francisco
In a new research article, scientists at Chan Zuckerberg Biohub San Francisco (CZ Biohub SF) describe Omega, an open-source software tool that significantly advances the field of bioimage analysis. Omega harnesses the power of large language models (LLMs) to enable scientists to process and analyze biological images through natural language conversations rather than having to issue formal commands or write code.
Created by Loïc A. Royer and his team, and documented in a paper published June 10, 2024 in Nature Methods, Omega is a plug-in for napari, an open-source image viewer used worldwide in diverse scientific fields, especially in biomedical research.
Omega is tightly integrated with various LLMs, including OpenAI’s ChatGPT, allowing scientists to conduct sophisticated bioimage processing and analysis through intuitive, conversational interactions that issue all the required commands to the napari software in the background.
“Omega allows users to quickly generate and edit code to solve complex image processing tasks,” explained Royer, a senior group leader and director of imaging AI at CZ Biohub SF. “You still need to understand the basics of image analysis, but Omega significantly speeds up the process.”
By prioritizing ease of use, Omega democratizes bioimage analysis, as researchers without extensive programming skills can use Omega to perform high-level analyses, accelerating their workflow and generating greater insight into their imaging data. Furthermore, Omega’s collaborative features, such as a shared code editor, enhance teamwork and knowledge sharing within the scientific community, according to Royer.
Omega’s features include:
- Interactive image analysis: Users can instruct Omega to perform specific tasks, such as segmenting cell nuclei, counting objects, and generating detailed reports, all through simple conversational prompts.
- On-demand widget creation: Omega can create custom widgets tailored to user-defined tasks, facilitating specialized image filtering, transformations, and visualizations.
- An AI-augmented code editor: Omega includes an intelligent code editor that enhances code management with automatic commenting, error detection, and correction features.
- Multimodal capabilities: Beyond text, Omega can interpret visual data, integrating multiple data types to provide comprehensive image analysis.
With the recent rise of LLMs and other AI platforms, Royer has envisioned a future in which bioimaging researchers will engage in dialogues with the software tools they depend on, rather than simply “issuing commands.”
“The idea for Omega began with an invited perspective piece published in Nature Methods in 2023, in which I predicted that in the very near future bioimage analysis tasks will be solved through ‘conversations with the machine,’” said Royer. “Omega is a significant stride toward this vision.”
Members of the scientific community are already making use of Omega, which has been available for download from a GitHub repository since May 2023, with regular updates posted since then. “The feedback has been overwhelmingly positive — the software is being downloaded approximately 2,000 times per month — and it has inspired other researchers to explore similar ideas,” said Royer.
The source code for Omega is openly available on GitHub, inviting contributions and collaboration from the global research community. This openness ensures that Omega will continually evolve, Royer said, incorporating the latest technological advancements to meet the ever-changing needs of scientists worldwide.
Looking ahead, Royer and his team plan to not only maintain Omega, but to continue enhancing its capabilities. “We plan to make Omega smarter and more robust, and compatible with the best and latest LLMs as they appear,” he said.
Despite the striking recent advancements in LLMs, however, Royer emphasized that human expertise remains essential in research. “There will always be a need for human experts, but tools like Omega are going to remove bottlenecks, such as the need for coding skills to turn ideas into reality, and will dramatically increase productivity in science.”
For more information about Omega, or to access the source code, please visit the GitHub repository.
###
About the Chan Zuckerberg Biohub San Francisco
CZ Biohub San Francisco, part of the Chan Zuckerberg Biohub Network, is a nonprofit biomedical research center founded in 2016. CZ Biohub SF’s researchers, engineers, and data scientists, in collaboration with colleagues at our partner universities — Stanford University; the University of California, Berkeley; and the University of California, San Francisco — seek to understand the fundamental mechanisms underlying disease and develop new technologies that will lead to actionable diagnostics and effective therapies. Learn more at czbiohub.org/sf.
Journal
Nature Methods
DOI
10.1038/s41592-024-02310-w
Article Title
Omega — harnessing the power of large language models for bioimage analysis
Article Publication Date
10-Jun-2024




