Credit: University of Colorado Cancer Center
When University of Colorado Cancer Center researcher, Jing Hong Wang, MD, PhD, found more than 1,000 genetic translocations in her mouse model of B cell lymphoma, she assumed her lab had made a mistake. To rule out experimental technique as the cause of the way-more-than-expected genomic alterations, Wang's lab sequenced three different types of cells from "wildtype" mice – effectively the kind that might move into your garage in bad weather. Like the lymphoma cells before them, the cells from wildtype mice also had over a 1,000 translocations.
"We thought 'let's just do another practice'," says Wang, also an associate professor in the CU School of Medicine Department of Immunology & Microbiology.
For "practice", paper co-first author, Katherine Gowan, downloaded new mouse genomic data from the website of Wellcome Trust Sanger Institute outside Cambridge in the UK, one of the world's leading institutes for genetic research. Gowan is a researcher with the group of Kenneth Jones, PhD, co-director of the CU Cancer Center Bioinformatics Shared Resource.
"When we mapped the genome of this particular mouse strain against the mouse reference genome published by the National Center for Biotechnology Information, we found thousands of translocations, even more than our experimental model!" Wang says.
The problem was not their experimental mouse. The problem was not the quality of their data nor the computational algorithm they used to discover translocations. The problem, as reported in an article in the journal BMC Genomics, was that reference genomes are different for various mouse strains. Not all mice have the same DNA sequences in the same locations on their chromosomes — due to this genetic variation, the DNA sequences of one mouse strain may appear out of place when compared with the DNA sequences of any other mouse strain.
The goal of this research was to discover new translocations that could be driving lymphoma. These translocations – accidental genetic rearrangements in which a gene is snipped from one location and pasted into another, sometimes creating a "fusion gene" made from both – have been implicated in a range of cancers, for example ALK-positive lung cancer, which is driven by the translocation of the ALK gene, which fuses with the gene EML4. The question was whether a similar translocation might be to blame for a subset of lymphomas.
"Unfortunately, when we have so many events, the artifacts may mask our real events," says Wang, meaning that with thousands of translocations identified by next-generation sequencing, it was almost impossible to discover the "needle" of a potentially oncogenic translocation amid the "haystack" of identified translocations that were, in fact, only the unimportant, random differences between individual mouse genomes.
"Then we started to think about all these human cancer genomic studies," Wang says. "People use all this sequencing data to show genomic changes in human cancers, but what if these studies have similar comparison problems?"
First, Wang points out, this possible trap is irrelevant when analyzing a patient's cancer for any known genetic change. In the previous example of lung cancer, genomic testing (often using the technique of fluorescent in situ hybridization or FISH) can tell if a cell's chromosomes do or do not contain an ALK-EML4 fusion gene. But it is when searching for important differences between a human cancer cell and a healthy human cell that the genetic backgrounds of these cells may skew results – due to the randomness of repeats and gene polymorphisms and other unpredictable genetic variations, the differences between a cancerous and a healthy cell may be due to chance and not to the influence of the cancer at all.
Part of the problem is the small size of genetic "snips" used by today's next-generation sequencing technology. In "next-gen seq" the machine reads a test genome as many snips, each made up of 100 to 150 base pairs. Then the computational biologist fits these snips like puzzle pieces against a reference genome. When there is a match, the system puts the piece in place and thus, because it knows the makeup of the reference genome, can come to know the makeup of the test genome. Unfortunately, with 3 billion base pairs in the human genome, there may be many false matches for short, 100 base-pair snips. Technology is on the way to solve this problem, sequencing the genome in much longer snips (1,000 or more base pairs).
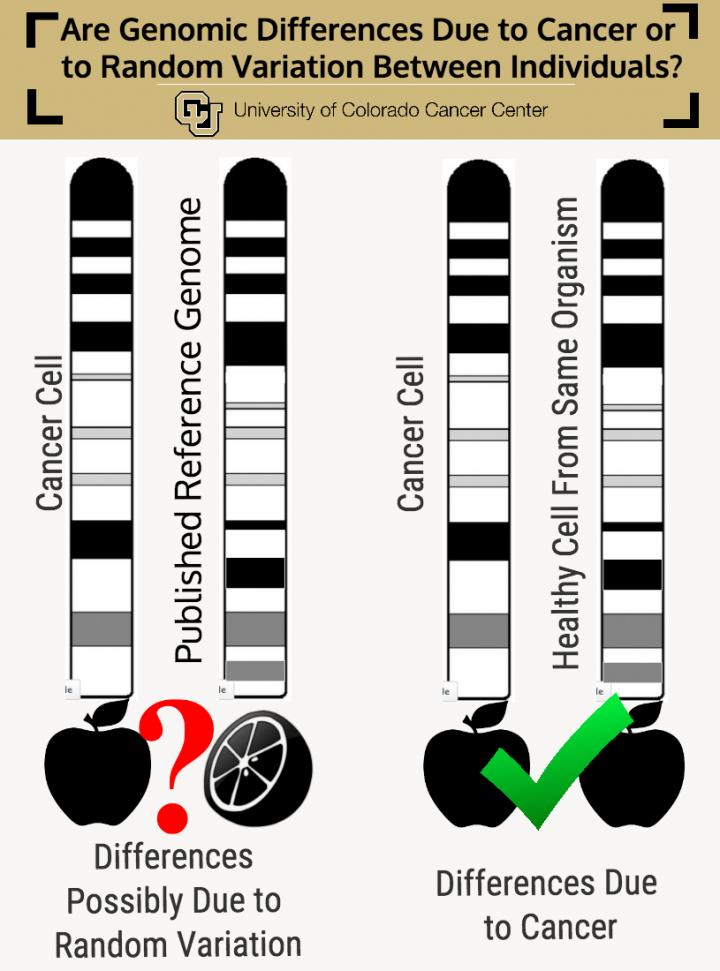
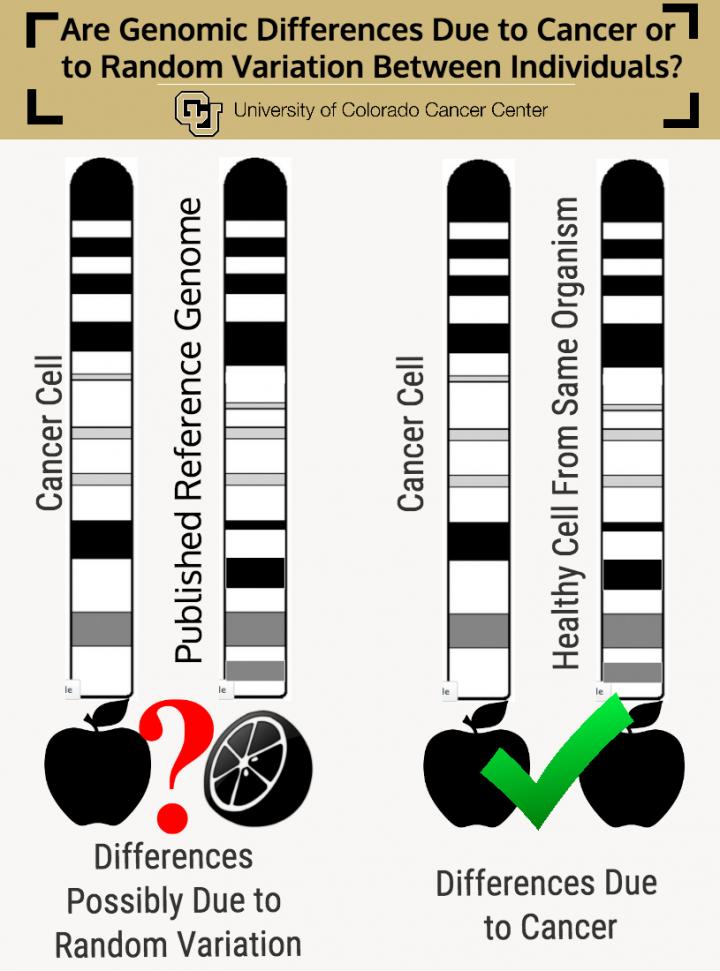
Until then, Wang suggests a possible fix: "We suggest considering not mapping your data to a reference genome, but to the genome of some cell from the same source that doesn't have cancer."
The paper calls this process "de novo assembly" — basically, instead of comparing a cancerous apple to a healthy orange, it is comparing a cancerous apple to a healthy apple.
"People should be their own control. Instead of working with the published, generic reference genome, we should work with two samples (control vs. cancer) from the same person," Wang says. "Only then can you really figure out what's going on in your cancer cell genome."
###
Media Contact
Garth Sundem
[email protected]
@CUAnschutz
http://www.ucdenver.edu