A new paper in the Journal of Chemical Information and Modeling provides definitive validation of PandaOmics, the cloud-based software platform from artificial intelligence (AI)-driven drug discovery company Insilico Medicine (“Insilico”) that is used to identify therapeutic targets and discover new biomarkers. PandaOmics is a critical component of Insilico’s end-to-end Pharma.AI platform, and uses proprietary AI algorithms to process vast quantities of diverse text and omics data and perform gene and pathway analysis and target predictions, all with a user-friendly interface.
Credit: Insilico Medicine
A new paper in the Journal of Chemical Information and Modeling provides definitive validation of PandaOmics, the cloud-based software platform from artificial intelligence (AI)-driven drug discovery company Insilico Medicine (“Insilico”) that is used to identify therapeutic targets and discover new biomarkers. PandaOmics is a critical component of Insilico’s end-to-end Pharma.AI platform, and uses proprietary AI algorithms to process vast quantities of diverse text and omics data and perform gene and pathway analysis and target predictions, all with a user-friendly interface.
“Target identification is a complex and critical part of the early drug discovery process,” says Insilico Medicine founder and co-CEO Alex Zhavoronkov, PhD. “So many drugs in development ultimately fail in clinical trials – a major drain on time and resources – due to poor efficacy. That, in turn, stems from choosing the wrong target.”
PandaOmics’ algorithm – iPANDA – performs pathway activation analysis, allowing users to understand which biological processes, such as autophagy or DNA replication, are implicated in a disease. It also highlights connections between genes, biological pathways, and metadata in the context of a particular disease of interest. Through meta-analysis, users can then aggregate multiple disease-relevant genetic data.
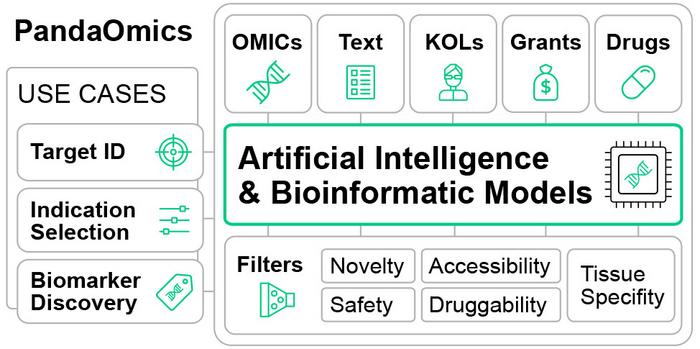
Data comes from numerous sources, including text data from scientific publications, grant applications, clinical trials, and omics data, which includes gene expression, genetics, proteomics, and methylation data. Within PandaOmics’ Target ID interface, there are 23 disease-specific models. Some of these are omics-based, relying on bioinformatics approaches and advanced machine learning and graph-based techniques, such as the expression score, which compares the differential expression of disease samples to paired normal control samples and the expression levels in disease-relevant tissues. The interface allows users to rank the resulting genes of interest based on specified criteria, which might include druggability by small molecules and therapeutic antibodies, safety considerations, novelty of the target, tissue-specific expression patterns, protein class, biological process involvement, availability of crystal structures, and the level of pharmaceutical development.
For users who are interested in looking at how a target might be relevant in multiple diseases, PandaOmics has an Indication Prioritization feature which is presented similarly to the Target ID heatmap. Diseases are conveniently grouped to align with the pipeline divisions of leading pharmaceutical enterprises and can be further categorized by therapeutic domains or specific tissue/organ systems. This prioritization feature relies on a repository of pre-calculated disease meta-analyses encompassing over 8,000 diseases, with a dedicated emphasis on more than 500 manually curated meta-analyses. PandaOmics also scores compounds and compares them to known targets or disease-associated genes, allowing for further prioritization.
PandaOmics also has a Knowledge Graph that utilizes advanced algorithms that draw on publications, clinical trials, and other data to provide a deeper understanding of the competitive landscape to underscore the value of a particular identified target. Users can easily interact with this graph using the tool’s ChatPandaGPT functionality, a large language model feature that provides relevant summaries and answers questions.
Insilico’s AI-powered next-generation robotics lab further enhances PandaOmics’ capabilities. As the robotics lab performs target and compound validation, its sequencing and phenotypic data are fed back into PandaOmics, enriching the dataset and enhancing the accuracy of target and biomarker prediction. In turn, PandaOmics’ insights help guide the design and selection of targets for further validation and testing.
In its fourth iteration, PandaOmics has been extensively validated in biomarker discovery and target identification across multiple therapeutic areas, including oncology, inflammation, and immunology. Successful case studies include the identification of potential biomarkers associated with androgenic alopecia, as well as with gallbladder cancer and smoke-induced lung cancer. PandaOmics has also successfully identified potential therapeutic targets for idiopathic pulmonary and kidney fibrosis, aging, glioblastoma multiforme, and head and neck squamous cell carcinoma. Insilico’s lead AI-designed drug candidate for idiopathic pulmonary fibrosis, designed for a PandaOmics-identified target, is now in Phase II trials with patients.
As it has been shown in the recent 2023 papers, PandaOmics identified CAMMK2, MARCKS, and p62 – targets successfully validated in Alzheimer’s disease cell models and KDM1A as a dual aging and oncology target. Insilico scientists worked with a consortium of researchers in 2022 to identify 28 potential therapeutic targets for ALS using PandaOmics, which were later validated in animal models.
“It has been very exciting to see the advances in this platform in just a few years,” says Petrina Kamya, PhD, Head of AI Platforms and President of Insilico Medicine Canada. “PandaOmics is truly a state-of-the-art tool for early drug discovery and works seamlessly with other emerging technologies – including AlphaFold and new methods for detecting protein phase separation – to further advance its capabilities.”
In 2023, Insilico published the successful application of Chemistry42, in combination with the AlphaFold protein structure prediction tool and PandaOmics, to identify a novel hit molecule for liver cancer against a novel target, CDK20, that lacked an experimental structure. In 2023, Insilico published findings with a research team at the University of Cambridge on using PandaOmics with the FuzDrop method for predicting protein phase separation (PPS) to identify PPS-prone, disease-associated proteins.
PandaOmics is part of an end-to-end suite, Pharma.AI, that includes Chemistry42 for small molecule drug design and inClinico for virtual predictions of clinical trial outcomes. This software suite has contributed to Insilico’s robust internal pipeline of 31 drugs in development for cancer, immunotherapy, fibrosis, IBD, and COVID-19, with 5 therapeutic small molecules in clinical stages.
About Insilico Medicine
Insilico Medicine, a global clinical-stage biotechnology company powered by generative AI, connects biology, chemistry, and clinical trial analysis using next-generation AI systems. The company has developed AI platforms that utilize deep generative models, reinforcement learning, transformers, and other modern machine learning techniques for novel target discovery and generating novel molecular structures with desired properties. Insilico Medicine is developing breakthrough solutions to discover and develop innovative drugs for cancer, fibrosis, immunity, central nervous system diseases, infectious diseases, autoimmune diseases, and aging-related diseases. www.insilico.com
Journal
Journal of Chemical Information and Modeling
DOI
10.1021/acs.jcim.3c01619
Article Title
PandaOmics: An AI-Driven Platform for Therapeutic Target and Biomarker Discovery
Article Publication Date
25-Feb-2024




