Credit: University of Washington
Living cells must constantly process information to keep track of the changing world around them and arrive at an appropriate response.
Through billions of years of trial and error, evolution has arrived at a mode of information processing at the cellular level. In the microchips that run our computers, information processing capabilities reduce data to unambiguous zeros and ones. In cells, it's not that simple. DNA, proteins, lipids and sugars are arranged in complex and compartmentalized structures.
But scientists — who want to harness the potential of cells as living computers that can respond to disease, efficiently produce biofuels or develop plant-based chemicals — don't want to wait for evolution to craft their desired cellular system.
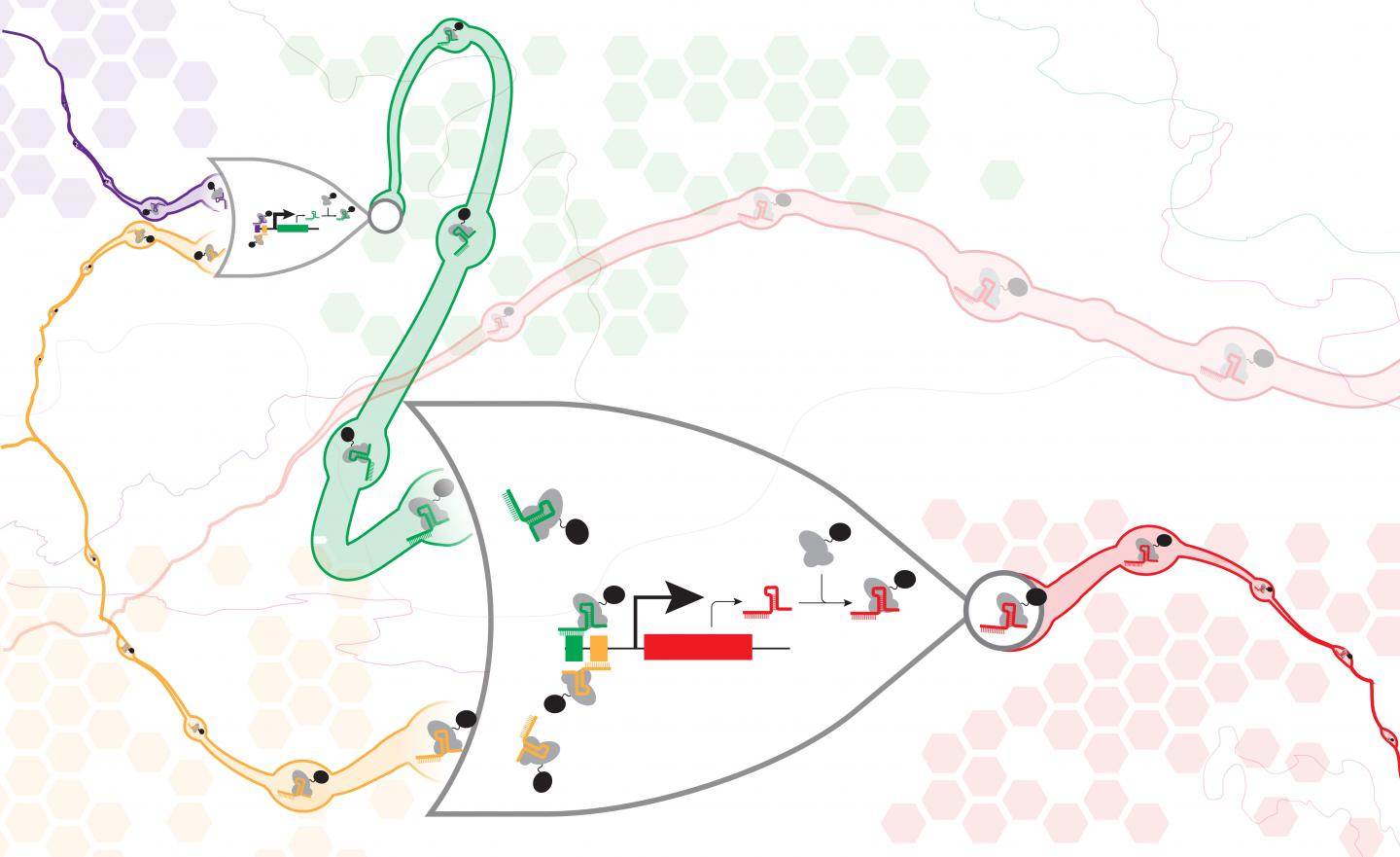
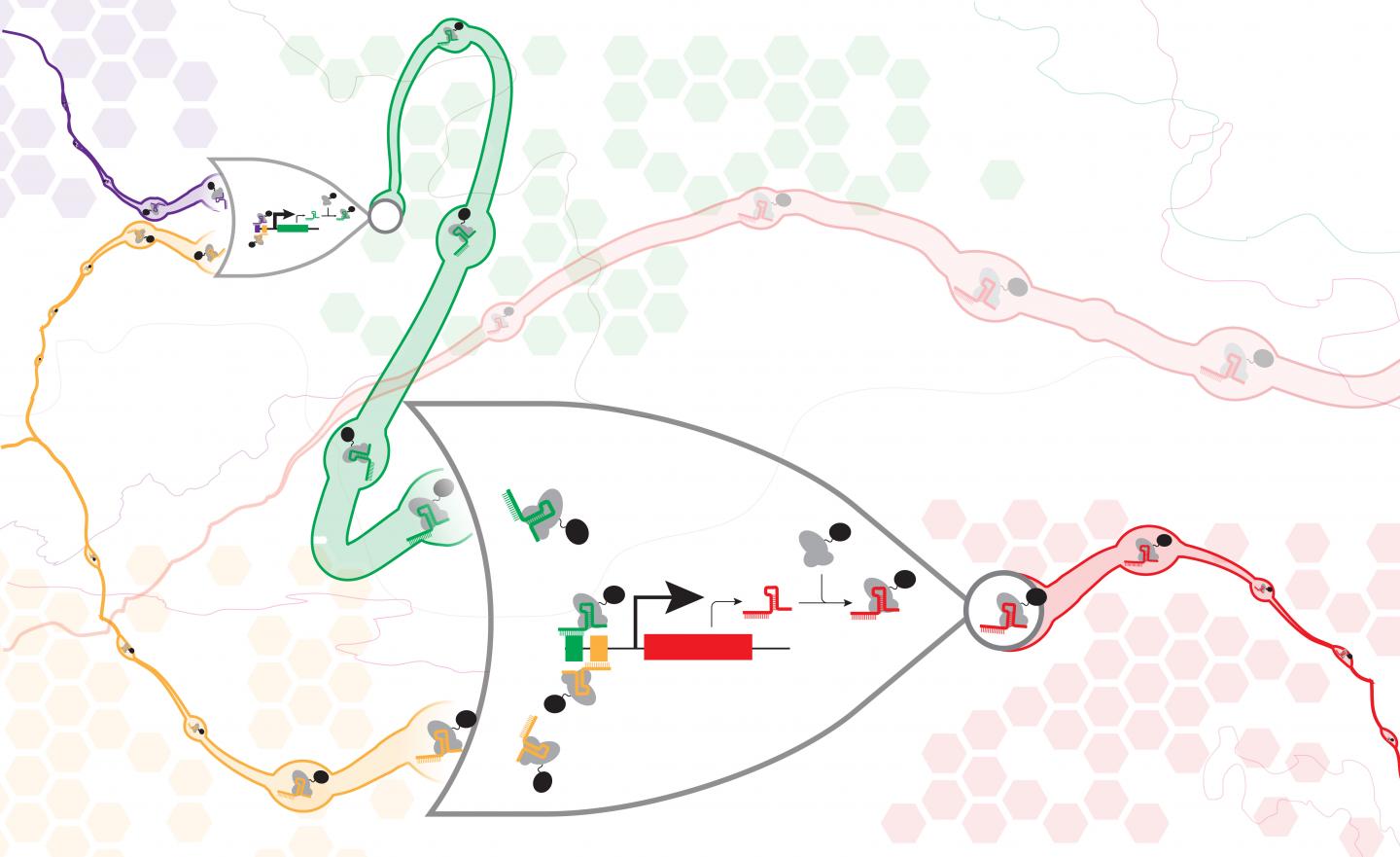
In a new paper published May 25 in Nature Communications, a team of UW synthetic biology researchers have demonstrated a new method for digital information processing in living cells, analogous to the logic gates used in electric circuits. They built a set of synthetic genes that function in cells like NOR gates, commonly used in electronics, which each take two inputs and only pass on a positive signal if both inputs are negative. NOR gates are functionally complete, meaning one can assemble them in different arrangements to make any kind of information processing circuit.
The UW engineers did all this using DNA instead of silicon and solder, and inside yeast cells instead of at an electronics workbench. The circuits the researchers built are the largest ever published to date in eurkaryotic cells, which, like human cells, contain a nucleus and other structures that enable complex behaviors.
"While implementing simple programs in cells will never rival the speed or accuracy of computation in silicon, genetic programs can interact with the cell's environment directly," said senior author and UW electrical engineering professor Eric Klavins. "For example, reprogrammed cells in a patient could make targeted, therapeutic decisions in the most relevant tissues, obviating the need for complex diagnostics and broad spectrum approaches to treatment."
Each cellular NOR gate consists of a gene with three programmable stretches of DNA — two to act as inputs, and one to be the output. The authors then took advantage of a relatively new technology known as CRISPR-Cas9 to target those specific DNA sequences inside a cell. The Cas9 protein acts like a molecular gatekeeper in the circuit, sitting on the DNA and determining if a particular gate will be active or not.
If a gate is active, it expresses a signal that directs the Cas9 to deactivate another gate within the circuit. In this way, the researchers can "wire" together the gates to create logical programs in the cell.
What sets the study apart from previous work, researchers said, is the scale and complexity of the circuits successfully assembled — which included up to seven NOR gates assembled in series or parallel.
At this size, circuits can begin to execute really useful behaviors by taking in information from different environmental sensors and performing calculations to decide on the correct response. Imagined applications include engineered immune cells that can sense and respond to cancer markers or cellular biosensors that can easily diagnose infectious disease in patient tissue.
These large DNA circuits inside cells are a major step toward an ability to program living cells, the researchers said. They provide a framework where logical programs can be easily implemented to control cellular function and state.
###
The research was funded by the Semiconductor Research Corporation and the National Science Foundation.
Co-authors include UW electrical engineering graduate student Miles Gander, bioengineering graduate student Justin Vrana, chemical engineering graduate student Willy Voje and chemical engineering professor James Carothers.
For more information, contact Eric Klavins at [email protected].
Media Contact
Jennifer Langston
[email protected]
206-543-2580
@UW
http://www.washington.edu/news/
############
Story Source: Materials provided by Scienmag